2358
A nested iteration artificial neural network approach for efficient high dimensional parameter estimation in 31P-MRF1CIBM Center for Biomedical Imaging, Lausanne, Switzerland, 2Animal Imaging and Technology, Ecole Polytechnique Federale de Lausanne (EPFL), Lausanne, Switzerland, 3Laboratory of functional and metabolic imaging, Ecole Polytechniqe Federale de Lausanne (EPFL), Lausanne, Switzerland
Synopsis
Keywords: MR Fingerprinting/Synthetic MR, Machine Learning/Artificial Intelligence, MRS, Phosporus, Creatine Kinase, MRF
A new magnetization transfer 31P Magnetic Resonance Fingerprinting (31P-MRF) technique is emerging to measure the creatine kinase (CK) chemical exchange rate kCK. The inherent obstacle of the exponential growth in the size of dictionaries with the number of free parameters, was overcome by introducing the nested iteration interpolation method (NIIM). To further reduce the processing time and cope with a nonlinear behaviour, we employed an artificial neuron network (ANN), instead of an interpolation method (IPM) as in the original approach. The nested iteration ANN method (NIAN) is compared with NIIM using simulation data and in vivo 31P-MRF data.Introduction
A new magnetization transfer (MT) phosphorus Magnetic Resonance Fingerprinting (31P-MRF) [1] technique is emerging to measure the creatine kinase (CK) chemical exchange rate kCK, which is key in energy buffering and transport. This framework can largely reduce the scanning time compared to the conventional MT-31P spectroscopy methods. The inherent obstacle of the exponential growth in the size of dictionaries with the number of free parameters, was overcome by introducing the nested iteration interpolation method (NIIM) [1]. NIIM showed a great efficiency in both computational and time to estimate 6 free parameters. However, multiple nested iteration paths still need to be computed to perform the linear interpolation. To further reduce the processing time and cope with a nonlinear behaviour, we employed an artificial neural network (ANN), instead of an interpolation method (IPM) as in the original approach. Therefore, this study aims to evaluate the performance of the nested iteration ANN method (NIAN) and compare it with NIIM using simulation data and in vivo 31P-MRF data.Methods
NIIM was recently introduced by Widmaier et al. [1] to overcome the exponential growth of memory and computation time for the generation of dictionaries in the MRF framework [2,3] for high dimensional multi parameter estimations. Like NIIM, the pattern matching process is split into several nested iterations instead of matching all the parameters simultaneously. A schematic framework is shown in Figure 1. The nested iteration is looped Nl=5 times, each time with a different kCK starting value kCK 0=[0.35,0.2,0.5,0.3,0.4]s-1. All other parameters in an iterative step are either free (objects of the estimation) or fixed. Parameters are fixed to literature values or estimated values in previous steps. Successively all parameters are estimated. In the last iterative step, all parameters are fixed to estimated values and kCKn is the objective of the estimation. The other 5 parameters are the longitudinal relaxation rates T1ATP,n and T1PCr,n, the off resonance foffn, the B1 factor fB1n and the concentration ratio Crn=M0PCr/M0ATP. The 6 estimates in the nth loop are biased, dependent on the starting value kCK0,n. These biased values are then used to find a corrected estimation by either applying the interpolation method (IPM) or an ANN.The ANN holds four hidden layers (Nin-50-25-15 neurons). Where Nin is the number of input features depending on the number of loops used. A simulated dataset was generated, with 1000 samples with randomly initialized parameters (described above) and added gaussian noise (signal-to-noise-ratio SNR=[0,3,6,9,12,100]dB). It was split into training, validation and test datasets [70/20/10] for simulation validation. Estimations were evaluated for different Nin by choosing the Nl=[1,3,5]. In vivo data acquisition is described in Widmaier et al. [1].
The estimation error was evaluated using the mean absolute percentage error and its standard deviation (STD) for the simulations. In-vivo the test-retest reproducibility was analysed with the coefficient of variance (CV).
Results
Figure 2 shows the comparison of estimation errors for each parameter when applying IPM or ANN in simulated datasets with various SNRs. Additionally, the performance of ANN is evaluated for different numbers of input features. Overall, the ANN shows a more accurate and robust estimation. The number of input features and nested iteration loops have no significant impact on the accuracy.Table 1 displays mean and standard deviation of estimated parameters from in vivo data over different acquisition times. The estimated values are consistent with those obtained by NIIM and EBIT [4].
The reproducibility is evaluated in figure 3. The CVs of parameters estimated by IPM and ANNs trained under different noise conditions are displayed over the acquisition time. The noise level of the training data does not significantly influence the CVs. The ANN shows a lower CV compared to IPM for all parameters at full acquisition length. For short acquisitions (<9 min) IPM has a lower CV for kCK.
Discussion and Conclusion
We demonstrate with simulation data, that the NIAN is more accurate and robust in a shorter processing time relative to the NIIM. The exploratory work proved that NIAN needs data just from a single nested iteration path, therefore, processing time and memory can be reduced by 80%. In-vivo human brain data at 7T showed the estimated values are consistent with those reported in the literature and comparable to those obtained with the NIIM and the efficient 31P band inversion transfer approach (EBIT) [4]. Overall, the ANN also possesses superior reproducibility on subjects over the IPM through a test-retest experiment. We conclude, that applying NIAN the in the MT-31P-MRF pattern matching process is feasible and has great potential in a fast and quantitative measurement of metabolic reactions.Acknowledgements
This work was supported by the Swiss National Science Foundation (grants n° 320030_189064). We acknowledge access to the facilities and expertise of the CIBM Center for Biomedical Imaging, a Swiss research center of excellence founded and supported by Lausanne University Hospital (CHUV), University of Lausanne (UNIL), Ecole polytechnique fédérale de Lausanne (EPFL), University of Geneva (UNIGE) and Geneva University Hospitals (HUG).”References
[1] M. Widmaier, S.-I. Lim, D. Wenz, and L. Xin, “Fast in vivo assay of creatine kinase in human brain by 31P magnetic resonance fingerprinting,” Jun. 2022. https://www.researchsquare.com/article/rs-1708658/v1
[2] Ma D, Gulani V, Seiberlich N, et al. Magnetic resonance fingerprinting. Nature. 2013;495(7440):187-192. doi:10.1038/nature11971
[3] Wang CY, Liu Y, Huang S, Griswold MA, Seiberlich N, Yu X. 31P magnetic resonance fingerprinting for rapid quantification of creatine kinase reaction rate in vivo. NMR in Biomedicine. 2017;30(12):e3786. doi:10.1002/nbm.3786
[4] Ren J, Sherry AD, Malloy CR. Efficient 31P band inversion transfer approach for measuring creatine kinase activity, ATP synthesis, and molecular dynamics in the human brain at 7 T. Magnetic Resonance in Medicine. 2017;78(5):1657-1666. doi:10.1002/mrm.26560
Figures
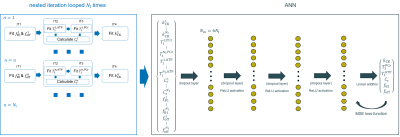
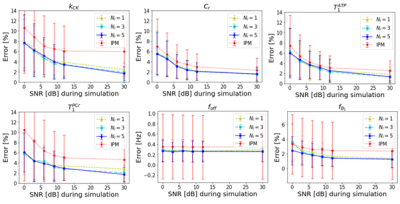
Figure 2: Mean and standard error comparison between IPM and ANN bias correction method over SNR [dB] for each parameter. For the ANN different numbers of nested iteration path loops Nl are displayed.
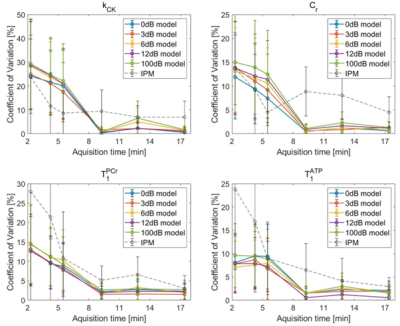
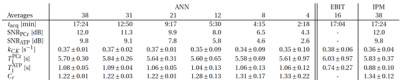
Table 1: In-vivo parameter estimations for different acquisition times. As reference results from EBIT and IPM are showed.