2265
Compressed SENSE AI enhanced mDixon-Quant MR Imaging in the Lumber Spine Study1Department of Medical Imaging, the Third Xiangya Hospital, Central South University, Changsha, China, 2Philips Healthcare, Guangzhou, China
Synopsis
Keywords: MSK, Fat, CS AI, Bone Marrow Fat, mDIXON-quant
Compressed SENSE (CS) has been suggested to speed up MRI acquisition in clinical studies, while reducing artefacts and improving image quality. To date, the optimal acceleration factor (AF) for Compressed SENSE AI (CS AI) versus conventional compressed SENSE (CS) on lumber spine images remains unclear. In this study, the impact of CS AI technique with different acceleration factors compared with conventional CS on the utility of measuring lumber spine fat was investigated. Results of this study showed that CS AI not only shortened MRI acquisition time, but also ensured image quality, as well as clinical diagnostic accuracy and clinical throughput.Introduction
The bone marrow is one of the largest tissues in human body, which contains adipocytes, hematopoietic stem cells and mesenchymal stem cells responsible to produce bone cells and adipocytes. Studies have shown that increased bone marrow adiposity is associated not only with reduced bone mass, increased risk of fracture and osteoporosis, but also with diabetes, lack of nervous appetite and bone unloading[1]. Increased bone marrow adiposity is now a major public health concern[2].Recently, Compressed SENSE AI has been proved valuable in speeding up acquisition time and ensuring image quality, which attracts increasing interest from researchers. A key foundation of this approach is based on deep learning, which learns image structures and content information via Artificial Neural Networks like Convolutional Neural Networks (CNN) and Generative Adversarial Networks (GAN), etc., to generate a priori information for under-sampling image reconstruction, shortening acquisition time by reducing the number of sampling points for image reconstruction, and finally recovering the k-space under-sampled data using 3D reconstruction methods to ensure image quality[3-5]. What’s more, compressed SENSE has the advantage of reducing acquisition time, which makes it widely used clinically, and CS AI based on deep learning sampling and reconstruction scheme may provide a new technique for next generation MR Imaging. So far, it is not clear about the accuracy of different acceleration factors of CS AI for measuring bone marrow fat content and the quality of accelerated liver MR images.
The aim of this study was to estimate outcomes of CS AI and CS and choose the appropriate acceleration factors, in order to shorten lumber spine MR acquisition time and improve patient cooperation, while ensuring clinical diagnostic accuracy and reliability, when quantify the fat component within human lumber spine.
Methods
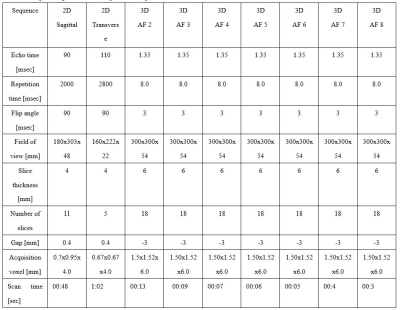
MRI Data acquisition and Preprocessing: 3D mDixon-Quant sequence measurements were obtained with hepatic MR imaging by using a Philips 3T scanner (Ingenia 3.0 ELITION, Philips Healthcare, Best, NL). The specific scanning protocols are shown in Table 1. A vendor-neutral postprocessing platform (Philips, Philips Healthcare) was used to draw regions of interest (ROIs). The image quality was evaluated via signal-to-noise (SNR) and contrast-to-noise ratios (CNR), which were computed as: SNR=SNR vertebrae/SD spinal cord, CNR=(SNR vertebrae - SNR spinal cord)/SD spinal cord. MR signal intensity of these data was measured within each ROI, respectively.Statistical analysis: The statistical analysis was performed with SPSS 26 software (SPSS, Chicago, IL, USA). The Wilcoxon signed-rank test was used to assess differences of image quality between CS and CS AI protocols. Cohen’s kappa was used to assess the consistency between measurements from two different observers,who scored image quality according to subjective criteria on the Likert scale. All statistical tests were defined as two-tailed, and p < 0.05 was considered as statistically significant.
Results
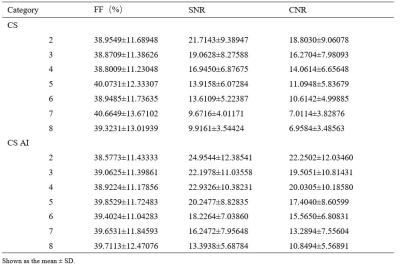
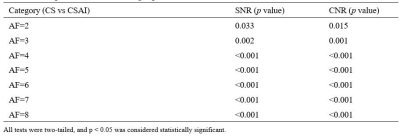
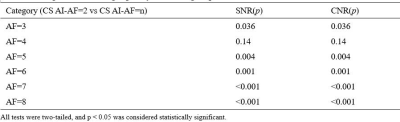
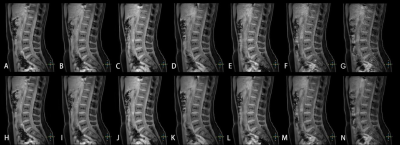
The consistency of measurement and subjective scores between the two observers was reasonable (ICC 0.679 to 0. 767, Kappa 0.723). Subjective scores from Observer 1 were selected for subsequent analysis.The Wilcoxon test was used to compare the difference between 3D mDixon-Quant sequences collaborated with different CS and CS AI protocols, while the lumbar vertebral Fat Fraction (FF) values and their image quality were also considered. Results showed that there was no statistically significant difference in the FF values of the lumbar vertebrae (p=0.998), but there was a statistically significant difference in SNR and CNR between CS AI and CS groups (p<0.001, p<0.001) (Table 2). Among them, there showed statistically differences in SNR and CNR among CS AI 2 groups, CS AI 3 groups, CS AI 4 groups, CS AI 5 groups, CS AI 6 groups, CS AI 7 groups, CS AI 8 groups(p<0.05), while groups like CS AI 2 and CS AI 4 groups showing no statistically differences (Table 3,4). Meanwhile, the scan times were further compared among all the groups, and the results showed that when the CSAI AF were 3, 4, 5, 6, 7 and 8, the scan times were 30.8%, 46.2%, 53.8%, 61.5%,69.2 % and 76.9% longer than when the CSAI AF was set as 2, respectively. Two physicians who scored the image quality found that as the AF increased, CS sequence image noise increased, too, and CS AI showed some image blurring (Figure 1).
Discussion & Conclusion
To our knowledge, this is the first study of CS AI accelerating 3D mDixon-quant sequence to quantify lumbar fat content and to further explore the effect of different acceleration factors on image quality. In this study, we found that the acquisition time became shorter when higher acceleration factors were used. Although the imaging quality did not increase with higher acceleration factors was set, an acceleration factor to some extend did not only shorten the acquisition time, but also ensure the image quality and proper clinical diagnosis. Among the different acceleration factors, although the acceleration factors of 7 and 8 are more effective in reducing the time, the image quality was severely degraded, and combined with the reduction in time and image quality when AF was set as 3, CS AI sequence showed the optimal protocol in this scenario, where CS tech failed. Findings within this study will help the clinician to observe subtle anatomical structures by using CS AI.Acknowledgements
None.References
[1] DEVLIN M J, ROSEN C J. The bone-fat interface: basic and clinical implications of marrow adiposity [J]. Lancet Diabetes Endocrinol, 2015, 3(2): 141-7.
[2] SEBO Z L, RENDINA-RUEDY E, ABLES G P, et al. Bone Marrow Adiposity: Basic and Clinical Implications [J]. Endocr Rev, 2019, 40(5): 1187-206.
[3] JOHNSON P M, RECHT M P, KNOLL F. Improving the Speed of MRI with Artificial Intelligence [J]. Semin Musculoskelet Radiol, 2020, 24(1): 12-20.
[4] ARSHAD M, QURESHI M, INAM O, et al. Transfer learning in deep neural network based under-sampled MR image reconstruction [J]. Magn Reson Imaging, 2021, 76(96-107.
[5] AKçAKAYA M, MOELLER S, WEINGäRTNER S, et al. Scan-specific robust artificial-neural-networks for k-space interpolation (RAKI) reconstruction: Database-free deep learning for fast imaging [J]. Magn Reson Med, 2019, 81(1): 439-53.
Figures