2235
A clinically applicable deep-learning model for automatic detection of focal liver lesions on Gd-DTPA-enhanced MRI1Department of Radiology, Beijing Friendship Hospital, Capital Medical University, Beijing, China, 2Shukun (Beijing) Technology Co., Ltd, Beijing, China
Synopsis
Keywords: Liver, Liver
Accurate detection of focal liver lesions in Gd-DTPA-enhanced magnetic resonance imaging (MRI) necessitates a high level of skill and experience. This task is typically performed by radiologists through visual inspection, which is time-consuming, labor-intensive, and subject to intra- and inter-observer variation. Convolutional neural networks (CNNs) have demonstrated significant potential in detecting lesions on medical imaging. Our study presents a unified multi-sequence lesion detector model for automatically detecting focal liver lesions on Gd-DTPA-enhanced MR images to aid in treatment decision-making.Introduction
Magnetic resonance imaging (MRI) has become the essential imaging examination to detect and diagnose focal liver lesions (FLLs) due to the detailed and comprehensive information provided1. However the accurate detection of lesions requires a great deal of skill and experience. A few small lesions, particularly those near the liver's margin or lesions with similar signal intensity to the surrounding liver parenchyma, are easily missed, especially by residents. This task is typically performed by radiologists through visual inspection, which is time-consuming, labor-intensive, and subject to intra- and inter-observer variation. Convolutional neural networks (CNNs) are the most widely used deep learning architecture in the field of medical image analysis2. It has demonstrated significant potential in detecting lesions on medical imaging3-6. The purpose of this study was to develop a unified multi-sequence lesion detector (MSLD) based on Gd-DTPA-enhanced MRI that includes unenhanced and enhanced images to automatically detect FLLs.Methods
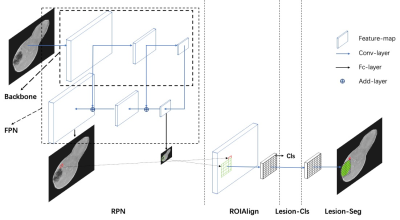
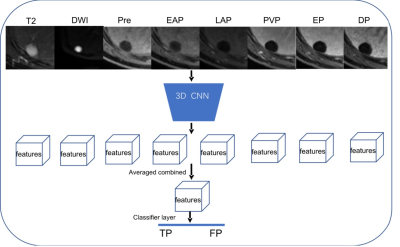
To train and validate the model, we respectively collected 3050 patients with multiphase DCE MR images from 4 hospitals. The unenhanced sequences included pre-contrast T1WI, T2WI, DWI (b value: 600 s/mm2@1.5T; 800s/mm2@3.0T) and ADC. The Gd-DTPA-enhanced phases included late arterial phase, portal venous phase and delayed phase. An external test cohort of images from 209 patients was included to further verify the model's generalization capability. The ground-truth was created by two radiologists with more than 5 years of experience who used dedicated software to draw bounding boxes around each FLL they detected on unenhanced and enhanced sequences. Disagreements were settled by majority vote with the help of a third radiologist with 25 years of experience.The first sub-task is to detect the lesions independently in each sequence. The MRI sequences are divided into four groups as pre-contrast T1WI, postcontrast T1WI, T2WI and DWI. Given the target sequence, the single lesion detector (SLD) produces a set of candidate lesion locations (denoted as bounding boxes) as outputs. SLD is built on Mask RCNN6, 7, an object detection framework proposed in the field of 2D natural images. We then adapt the network from 2D to 3D. The framework had four parts: region proposal network (RPN), ROI align, lesion classification (Lesion_Cls) and segmentation (Lesion_Seg) module (Figure 1). Furthermore, we used a receptive field (RF) adaptation model to improve the adaptive perception capability at the neuron-level. The second sub-task is to reduce false alarms using a false positive reduce (FPR) module. MSLD produces a set of lesions, each of which is associated with a set of bounding boxes from different sequences. The detection results of SLD have a branch of false alarm lesions that originate from the artifacts. FPR uses the appearance of the focal patch from multiple sequence to find the artifacts out. Figure 2 depicts the model architecture. A 3D-CNN module is used to extract image features of each crop as output, the features are averaged combined and feeding to the classifier layer and produce the binary prediction.Results
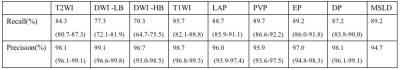
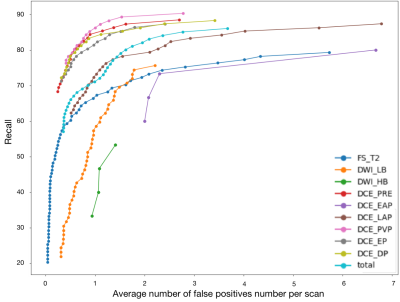
In the external cohort which was to test the model independently, the lesion-wise sensitivity on each single MRI sequence was listed in the Table 1. The FROC of each single MRI sequence was shown in the Figure 3. The recall of the MSLD model to detect lesions in portal phase was the highest, reaching 89.7%, while DWI with high b values was the lowest, reaching 70.3%. The lesion-wise sensitivity on the multiple MRI sequences was 89.2% (608/681), defined as the proportion of lesions detected on at least one of the multiple MRI sequences by the MSLD model among all lesions. The outcome is similar regardless of the manufacturer, Tesla, liver parenchyma, or malignant or benign lesions.Discussion
In this study, we developed an MSLD model to detect FLLs on Gd-DTPA-enhanced MRI images, including unenhanced and enhanced images, which is a basic but tricky problem. The modality diversity is one of the most difficult aspects of processing the family of MRI sequences listed in the acquisition protocol as a whole. The thickness of T2WI sequences can be 3~4 times that of T1WI sequences, making spatial normalization difficult. The appearance of the same tissue, such as a lesion or a vessel, in postcontrast T1WI, precontrast T1WI, T2WI, and DWI may differ. The variety of appearances could confuse a single detector. To adapt the diversity, we build SLD for each individual sequence. The issue of artifacts in MRI imaging is widespread and cannot be ignored. Because the majority of the artifacts appear in only a few sequences, the FPR model uses the other sequences to differentiate the artifacts from the lesions.There are a few limitations on this study. First, the model was trained and validated using retrospective data. Thus, more research and validation may be needed before such a system can be deployed and relied on in clinical practice. Secondly, the amount of data included in the external cohort was small, and the lesion type and liver background were not classified in detail. To check the model's robustness, more types of data must be added in the future.
Conclusion
Our study presents a unified multi-sequence lesion detector model for automatically detecting focal liver lesions on Gd-DTPA-enhanced MR images to aid in treatment decision-making.Acknowledgements
NoReferences
1. Matos A P, Velloni F, Ramalho M, et al. Focal liver lesions: Practical magnetic resonance imaging approach[J]. World J Hepatol, 2015, 7(16): 1987-2008.
2. Litjens G, Kooi T, Bejnordi B E, et al. A survey on deep learning in medical image analysis[J]. Med Image Anal, 2017, 42: 60-88.
3. Shi Z, Miao C, Schoepf U J, et al. A clinically applicable deep-learning model for detecting intracranial aneurysm in computed tomography angiography images[J]. Nat Commun, 2020, 11(1): 6090.
4. Xu L, He Y, Luo N, et al. Diagnostic Accuracy and Generalizability of a Deep Learning-Based Fully Automated Algorithm for Coronary Artery Stenosis Detection on CCTA: A Multi-Centre Registry Study[J]. Front Cardiovasc Med, 2021, 8: 707508.
5. Xue J, Wang B, Ming Y, et al. Deep learning-based detection and segmentation-assisted management of brain metastases[J]. Neuro Oncol, 2020, 22(4): 505-514.
6. Fu F, Wei J, Zhang M, et al. Rapid vessel segmentation and reconstruction of head and neck angiograms using 3D convolutional neural network[J]. Nat Commun, 2020, 11(1): 4829.
7. He K, Gkioxari G, Dollar P, et al. Mask R-CNN[J]. IEEE Trans Pattern Anal Mach Intell, 2020, 42(2): 386-397.
Figures