2216
To trigger or not to trigger – removing cardiac triggering in diffusion MRI of the cervical spinal cord saves time without sacrificing quality1Vanderbilt University Medical Center, Nashville, TN, United States, 2Polytechnique Montreal, Montreal, QC, Canada, 3Vanderbilt University, Nashville, TN, United States
Synopsis
Keywords: Data Acquisition, Spinal Cord
Cardiac triggering is commonly used in diffusion MRI protocols of the cervical spinal cord to reduce cardiac-related motion. However, this dramatically increases scan time and can limit high angular resolution multi-shell experiments. We test whether advances in preprocessing motion correction and fitting procedures may overcome cardiac-related motion artifacts. We find that removing cardiac triggering regains significant scan time with no increase in prevalence of artifacts, while providing similar quantitative indices with comparable reproducibility. In summary, removing cardiac triggering for cervical spinal cord diffusion saves time without sacrificing image quality.Introduction
Diffusion MRI of the cervical spinal cord (SC) is particularly challenging due to substantial physiological motion, including cardiac and respiratory motion and cerebrospinal fluid pulsation, which cause translation in the superior-inferior and anterior-posterior directions and a possibly nonlinear compression/stretching of the cord [1, 2]. Motion artifacts may cause signal or slice-dropout artifacts, misalignment of diffusion images, and subsequent biases in diffusion quantification [1]. To mitigate this, it has become standard practice to utilize cardiac triggering to acquire data at a phase of the cardiac cycle where motion is minimal [3]. However, triggering can dramatically increase scan time, which can be prohibitive in a clinical setting or in a research setting where high angular resolution multi-shell experiments are desired. Towards this end, we designed an experiment to directly test differences with/without triggering and assess image quality, artifacts, change in quantitative parameters, and reproducibility. Finally, we examined whether time saved by removing triggering can be used to acquire more directions without loss of image quality.Methods
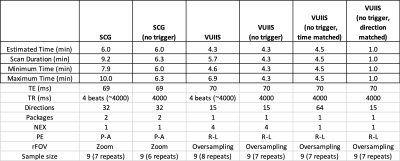
ProtocolOur experimental protocol includes 5 cervical spinal cord scans, each repeated twice within a scan session. We utilized the spinal cord generic protocol (SCG) [3, 4] and an in-house protocol (VUIIS) that both utilize triggering. Differences in the acquisition include number of directions, number of averages, and different reduced field of view techniques, among others. For each, we also acquire the same protocol with no triggering (SCG-nt; VUIIS-nt), with all other acquisition parameters matched (where the “estimated” scan duration is in theory equivalent). Next, we acquire a time-matched VUIIS protocol without triggering and without averaging, enabling the acquisition of 64 diffusion directions in an equivalent time (VUIIS-nt-time), from which we can select a subset of 15 directions to match the original protocol (VUIIS-nt-dir) which has an equivalent scan time of 1 minute.
Preprocessing
Preprocessing was done using a combination of MRTrix [5] for denoising, Spinal Cord Toolbox [6] for motion correction, segmentation, and metric extraction, and SCILPY for RESTORE robust tensor fitting and quality control metrics.
Comparisons
(1) Artifact prevalence was quantified through quantifying the % of slice signal dropouts, mean residuals from a tensor fit, physically implausible signals, and pulsation/misalignment derivation (see [5] for descriptions). (2) Change or bias in metrics due to triggering were quantified by calculating the tissue-probability weighted FA and MD in white and gray matter (C3-C4 level) with and without triggering. (3) Reproducibility was quantified by calculating a % Error from scan rescan measures of the same metrics. Significant differences between triggered and non-triggered protocols, as well as between direction- and time-matched protocols were determined using the nonparametric Wilcoxon sign-rank test with multiple comparison correction.
Results
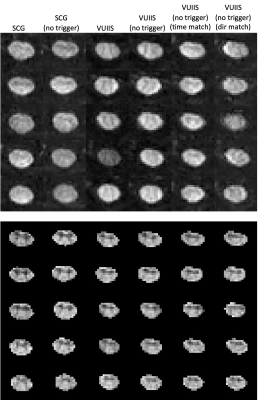
Table 1 shows the estimated and true scan durations of each protocol. While the estimated scan durations are matched, the true duration can be increased on average 46% and 32% with triggering (for SCG and VUIIS protocols, respectively).Figure 1 shows 5 middle slices for a single diffusion image and the derived FA map for an example subject for each acquisition protocol. Visually, very little differences are observed between trigger/non-triggering, and similar results are observed with the 1-minute reduced protocol (Figure 1, column 6).
Figure 2 plots quality control measures and shows that there is not a significant increase in any artifacts, residuals, or misalignment when removing triggering. However, there are increased residuals with the time-matched 64 direction sequence, likely due to the decreased SNR from decreased averaging.
Figure 3 shows that quantitative DTI values in white and gray matter are not statistically different when removing triggering from the acquisition protocol.
Figure 4 confirms that the scan-rescan reproducibility of derived indices does not significantly increase due to the removal of triggering, and in fact decreases in many cases (although the decrease is not statistically significant).
Discussion
It is common to utilize cardiac triggering in both clinical and research investigations of diffusion MRI in the cervical spinal cord. However, this detrimentally increases scan duration, which can negatively impact and limit studies requiring more diffusion directions or higher b-value data. Here, we show that when triggering is foregone, images are qualitatively similar, do not have increased prevalence of artifacts, result in similar diffusion tensor indices, and have similar reproducibility. When triggering is removed, very short acquisitions are possible, which are also qualitatively and quantitatively similar to triggered sequences.Acknowledgements
NIH K01EB030039 (KO), K01EB032898 (KS), 5R01NS109114 (SS), 5R01NS117816 (SS) and 5R01NS104149 (SS), R01EB017230 (BL)References
1. Morozov, D., et al., Effect of cardiac-related translational motion in diffusion MRI of the spinal cord.Magn Reson Imaging, 2018. 50: p. 119-124.
2. Winklhofer, S., et al., Spinal cord motion: influence of respiration and cardiac cycle. Rofo, 2014. 186(11): p. 1016-21.
3. Cohen-Adad, J., et al., Generic acquisition protocol for quantitative MRI of the spinal cord. Nat Protoc, 2021.
4. Cohen-Adad, J., et al., Open-access quantitative MRI data of the spinal cord and reproducibility across participants, sites and manufacturers. Sci Data, 2021. 8(1): p. 219.
5. Tournier, J.D., et al., MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. Neuroimage, 2019. 202: p. 116137.
6. De Leener, B., et al., SCT: Spinal Cord Toolbox, an open-source software for processing spinal cord MRI data. Neuroimage, 2017. 145(Pt A): p. 24-43.
7. Tournier, J.D., S. Mori, and A. Leemans, Diffusion tensor imaging and beyond. Magn Reson Med, 2011. 65(6): p. 1532-56.
Figures