2215
Assessment of neurochemistry in human-derived cerebral organoids using high-resolution magic angle spinning NMR1Physical Sciences, Sunnybrook Research Institute, Toronto, ON, Canada, 2Medical Biophysics, University of Toronto, Toronto, ON, Canada, 3Biological Sciences, Sunnybrook Research Institute, Toronto, ON, Canada, 4Chemistry, University of Toronto, Toronto, ON, Canada, 5University of Birmingham, Birmingham, United Kingdom
Synopsis
Keywords: Data Acquisition, Tissue Characterization, NMR
Cerebral organoids are self-organizing three-dimensional clusters of brain tissue, derived from human pluripotent stem cells. A new and rapidly advancing technology, cerebral organoids serve as an important model system for human brain research. In this study, we used high-resolution magic angle spinning (HR-MAS) NMR to characterize the neurochemical profile of ~100-day old cerebral organoids. High-quality spectra were obtained with excellent spectral resolution. More than 17 metabolites were detected, including many resonances commonly observed in the brain in vivo (e.g. choline, creatine, glutamate, GABA, etc.). Notably, NAA was absent. Future work will assess cerebral organoids at later stages of maturity.Introduction
Human pluripotent stem cell (hPSC)-derived cerebral organoids (COs) represent a breakthrough in neuroscience that allow researchers to study individualized human brain tissue in detail, thus serving as an important model system for human brain research [1-3]. Derived from human cells, COs take on many of the genetic , epigenetic and phenotypic characteristics of the individual donor, suggesting a potential role for COs in personalized medicine [3-8]. In this study, our goal was to develop high-resolution magic angle spinning (HR-MAS) NMR spectroscopy [9] for assessing neurochemical concentrations in hPSC-derived COs.Methods
Human-derived cerebral organoids: Human embryonic stem cells (hESCs) were used to derive 3D COs using a modified version of an established protocol [10], as illustrated in Figure 1. Briefly, hESCs were plated in embryoid body (EB) seeding medium. After 5 days, newly formed EBs were transferred to plates containing StemCell Tech CO induction medium. On day 9, EBs were embedded in Matrigel and deposited into plates containing StemCell Tech expansion medium. From day 5 to day 13, media was supplemented with 1 μM CHIR-99021 (StemCell Tech; #72052) and 1 μM SB-431542 (StemCell Tech; #72232). On day 13, embedded EBs were transferred to a spinning bioreactor (Spin Omega [10]) containing maturation medium in a 37°C incubator. COs were immunostained with neural markers to confirm their identity (Figure 1).NMR spectroscopy: High-resolution comprehensive multiphase (CMP) NMR [11] experiments were performed on a Bruker Avance III 500 MHz 1H spectrometer equipped with a 4 mm 1H-13C-15N (2H lock) CMP NMR probe (Bruker, Switzerland AG, Fällanden, Switzerland) fitted with a magic angle gradient. Two COs were scanned at 103 and 104 days of maturity, respectively. Before scanning, each CO was weighed and placed in a 4mm reduced volume (50 μl) Teflon rotor (HZ07213, Bruker) with D2O for locking. For the duration of each experiment, samples were maintained at 278K and spun at the magic angle with a spin rate of 2500 Hz to prevent the rupture of biological tissue [12], while positioning the spinning sidebands of water outside of the spectral window [13]. 1D 1H experiments were collected using 16484 time-domain points, 10,000 Hz spectral width, 8 dummy scans, and 512 total scans. Presaturation using an effective B1 field of 100 Hz was prepended to all 1D experiments. All 1D 1H spectra were collected using a TR of 11s (5 x T1 of longest relaxing peak) to eliminate T1 weighting. Diffusion editing experiments were performed to emphasize large components using a bipolar pulse pair longitudinal encode-decode (BPPLED) sequence as described previously [14]. 2D correlation spectroscopy (COSY) and 1H-13C heteronuclear single-quantum coherence (HSQC) experiments were also acquired to aid in the assignment of spectral peaks. Spectra were processed in MATLAB (Natick MA, USA) using the FID-A toolkit [15], and processed spectra were fitted using the Abfit software [16]. Metabolite peak heights were estimated with reference to an external reference of alanine with a concentration of 20mM and a volume of 30 μl.
Results
Figure 2 shows the 1D 1H spectra obtained from 103- and 104-day hESC-derived COs. In both scans, narrow linewidth of <2.5 Hz was achieved. More than 17 metabolites were detected, including glucose, lactate, creatine, glutamine, glutamate, alanine, ethanol, glycine, myo-inositol, hypotaurine, phosphocholine (PCh), glycerophosphocholine (GPC), choline, GABA, N-acetylasparate, acetate, valine and leucine. Figure 3 shows the ABfit results in one of the acquired spectra using. Some metabolites are fit very well, while for others the fit was poor, as evidenced by the large fit residuals. We are currently working on improving the model fitting and metabolite quantification.Discussion
We used HR-MAS NMR Spectroscopy to assess the neurochemical content of COs at 103 and 104 days of maturity. At both time points, we observed a detailed neurochemical profile consisting of at least 17 detectable metabolites, and excellent spectral quality. Many of the neurochemicals observed in COs are also observed in the human brain in vivo, including glutamate, glutamine, GABA, lactate, glucose, myo-inositol, creatine and cholines. However, differences exist as well. For example, strong resonances from hypotaurine and ethanol, are not normally detected in the human brain, were observed in COs. Moreover, N-acetylasparate, which is normally abundant in human brain in vivo, was only observed in very small quantities in the CO spectra. More research is required to understand the neurochemical differences between COs and in vivo human brain. One possibility is that COs at this stage exhibit a neurochemical pattern similar to human fetal brain development [17], in which NAA remains low until 22-24 weeks (154-161 days) of gestational age. Using immunohistology (see Figure 1) we hope to develop an understanding of the specific cellular changes that underlie neurochemical maturation. Moreover, since COs take on many of the genetic and phenotypic characteristics of the human donor, our eventual goal is to produce COs derived from human patients with various diseases (Alzheimer’s Disease, ALS, schizophrenia), and to assess disease-related neurochemical alterations.Conclusion
NMR allows assessment of neurochemistry in human-derived COs. At ~100 days of age, COs display a rich neurochemical profile. Future work will aim to assess neurochemical development as the COs mature, and in disease.Acknowledgements
This work is supported by the Canadian Institutes for Health Research (JN, AS and CS, Grant #: PJT-183715).
References
1. Chiaradia, I. and M.A. Lancaster, Brain organoids for the study of human neurobiology at the interface of in vitro and in vivo. Nat Neurosci, 2020. 23(12): p. 1496-1508.
2. Di Lullo, E. and A.R. Kriegstein, The use of brain organoids to investigate neural development and disease. Nat Rev Neurosci, 2017. 18(10): p. 573-584.
3. Slanzi, A., et al., In vitro Models of Neurodegenerative Diseases. Front Cell Dev Biol, 2020. 8: p. 328.
4. Zhao, J., et al., APOE4 exacerbates synapse loss and neurodegeneration in Alzheimer's disease patient iPSC-derived cerebral organoids. Nat Commun, 2020. 11(1): p. 5540.
5. Meyer, K., et al., REST and Neural Gene Network Dysregulation in iPSC Models of Alzheimer's Disease. Cell Rep, 2019. 26(5): p. 1112-1127 e9.
6. Yin, J. and A.M. VanDongen, Enhanced Neuronal Activity and Asynchronous Calcium Transients Revealed in a 3D Organoid Model of Alzheimer's Disease. ACS Biomater Sci Eng, 2021. 7(1): p. 254-264.
7. Zhao, J., et al., Apolipoprotein E regulates lipid metabolism and alpha-synuclein pathology in human iPSC-derived cerebral organoids. Acta Neuropathol, 2021.
8. Gonzalez, C., et al., Modeling amyloid beta and tau pathology in human cerebral organoids. Mol Psychiatry, 2018. 23(12): p. 2363-2374.
9. Beckonert, O., et al., High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nat Protoc, 2010. 5(6): p. 1019-32.
10. Qian, X., et al., Generation of human brain region-specific organoids using a miniaturized spinning bioreactor. Nat Protoc, 2018. 13(3): p. 565-580.
11. Fortier-McGill, B.E., et al., Comprehensive Multiphase (CMP) NMR Monitoring of the Structural Changes and Molecular Flux Within a Growing Seed. J Agric Food Chem, 2017. 65(32): p. 6779-6788.
12. Taylor, J.L., et al., High-resolution magic angle spinning proton NMR analysis of human prostate tissue with slow spinning rates. Magn Reson Med, 2003. 50(3): p. 627-32.
13. Mobarhan, Y.L., et al., Comprehensive multiphase NMR applied to a living organism. Chem Sci, 2016. 7(8): p. 4856-4866.
14. Courtier-Murias, D., et al., Comprehensive multiphase NMR spectroscopy: basic experimental approaches to differentiate phases in heterogeneous samples. J Magn Reson, 2012. 217: p. 61-76.
15. Simpson, R., et al., Advanced processing and simulation of MRS data using the FID appliance (FID-A)-An open source, MATLAB-based toolkit. Magn Reson Med, 2017. 77(1): p. 23-33.
16. Wilson, M., Adaptive baseline fitting for 1 H MR spectroscopy analysis. Magn Reson Med, 2021. 85(1): p. 13-29.
17. Girard, N., et al., MRS of normal and impaired fetal brain development. Eur J Radiol, 2006. 57(2): p. 217-25.
Figures
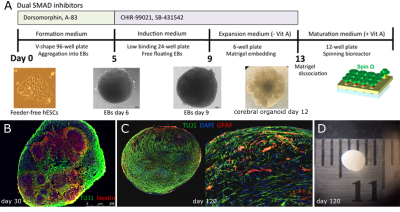
Generation of human-derived cerebral organoids (COs). (A) Small molecule strategy used to generate COs from Qian et al. 2018. (B-D) Sample COs showing expression of progenitor (Sox2, nestin in red) and neuronal (Tuj1 in green) markers after day 18 and 30. Day 120 COs grow to 3 mm in diameter.
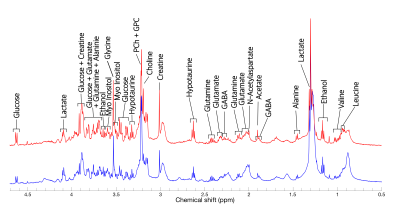
1D 1H NMR spectra acquired from human pluripotent stem cell-derived cerebral organoids at 103 (blue curve) and 104 (red curve) days of maturity. Excellent spectral quality is obtained, with linewidths of ~2.5 Hz. Good reproducibility is observed between organoids, and more than 17 metabolite resonances are detected.
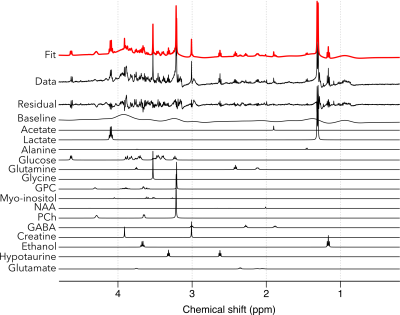
Linear combination model fit to one of the acquired cerebral organoid NMR spectra using the ABfit software. While some metabolite resonances are fit well, others are poorly fit, as evidenced by the fit residual. Work to improve the model fitting is currently in progress.