2209
Reduction of ADC bias with deep learning-based acceleration in diffusion-weighted MRI: A phantom validation study1Diagnostic and Interventional Radiology, Uniklinik RWTH Aachen University, Aachen, Germany, 2Philips Japan, Tokyo, Japan, 3Philips Healthcare, Best, Netherlands, 4Diagnostic and Interventional Radiology, University Hospital RWTH Aachen, Aachen, Germany, 5Philips GmbH Market DACH, Hamburg, Germany
Synopsis
Keywords: Image Reconstruction, Diffusion/other diffusion imaging techniques, Noise
Further acceleration of diffusion MRI in clinical examinations is desired but challenging mainly due to low signal and associated potential bias in the quantitative apparent diffusion coefficient (ADC) values. Artificial intelligence-based denoising and image reconstruction may provide a solution to address this challenge. We investigate and compare different image reconstruction methods, including conventional parallel imaging, compressed sensing, and a deep learning-based technique, in ADC accuracy and precision using a diffusion phantom with illustration of the principle in numeric simulation.Introduction
Diffusion-weighted MRI (DWI) plays an important role in clinical diagnosis, particularly in oncology. Further acceleration of DWI is clinically desired but challenging, mainly due to its low signal-to-noise ratio (SNR) especially at high b-values, and potential bias in the apparent diffusion coefficient (ADC) values associated with this. Typically, a sufficiently high number of b-factor averages needs to be acquired to prevent noise-related bias and loss of precision of ADC measurements1. Recently, deep learning-based reconstruction techniques have emerged, promising further acceleration of time-consuming sequences like DWI, while improving ADC robustness2,3. The purpose of this simulation-corroborated phantom study was to validate a deep-learning based image reconstruction method for DWI with comparison to conventional acceleration techniques.Methods
To investigate effects of noise on ADC bias and precision, nominal diffusion-weighted signals$$$M(b)=M_0·e^{-b·ADC}$$$ (Eq.1)
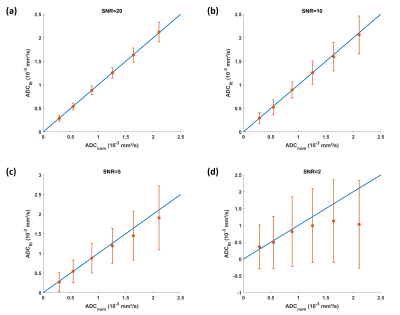
were simulated for a range of nominal ADC values (ADCnom) between 0.3·10-3 and 2.1·10-3 mm²/s, setting b=(50,400,1000)s/mm² and M0=1. Noisy signals with signal-to-noise ratios (SNR) between 2 and 20 were obtained by adding white Gaussian noise of different standard deviation (SD), where SNR=M0/SD1. Per ADCnom and SNR level, 200 noisy signals were fitted with Eq.1. Bias and precision were evaluated as mean and SD of the fitted ADC values.
For the actual phantom study, a diffusion phantom (CaliberMRI, Boulder, CO, USA) was measured in a 3.0T clinical MRI system (3T Elition X, Philips, Best, The Netherlands). A conventional single-shot echo-planar-imaging (EPI) DWI sequence with equidistant k-space undersampling for acceleration was employed. Key scan parameters include FOV=240mm, acq&rec matrix=128x128, slice thickness=1mm, FA=90°, TR=3500ms, TE=64ms, b-values=50,400 and 1000 s/mm².
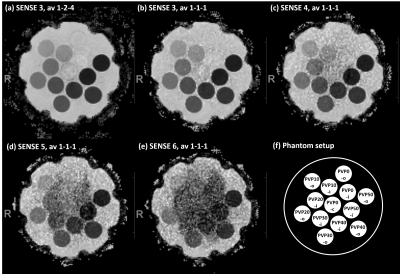
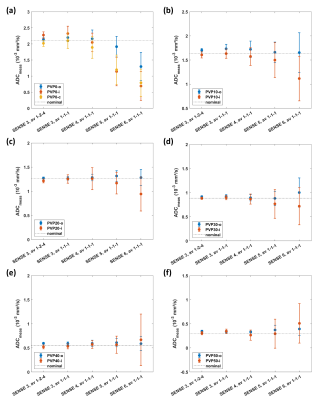
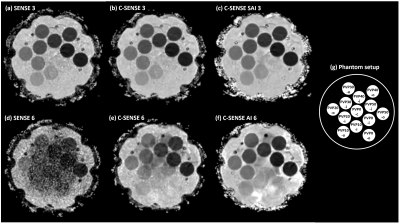
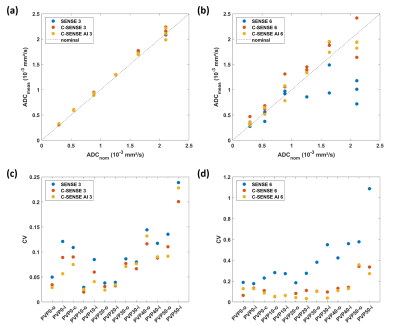
Measurements were performed in two setups. Setup 1 investigated the effect of SNR reduction on ADC. First, the numbers of b-factor averages for b=50-400-1000 s/mm2 were reduced from 1-2-4 to 1-1-1. Corresponding scan durations were 1min17s and 35s. Second, the acceleration factor was increased from 3 to 6, while the other parameters were fixed with b-value averages 1-1-1. Conventional SENSitivity Encoding (SENSE) reconstruction was employed. Setup 2 investigated the effect of reconstruction technique on ADC. To that purpose, two measurements were acquired with scan acceleration factors of 3 and 6, respectively; again, b-value averages 1-1-1 and other parameters were unchanged. Three reconstruction methods were applied, i.e., conventional SENSitivity Encoding (SENSE), compressed SENSE (C-SENSE) and compressed SENSE Artificial Intelligence (C-SENSE AI), all integrated to the vendor provided online reconstruction architecture. While C-SENSE employed compressed sensing principle including coil sensitivity information4,5, C-SENSE AI integrated a convolutional neural network (CNN) into the C-SENSE reconstruction based on the recently introduced Adaptive-CS-Net6, as a software prototype. For both setups, mean ADC and SD were measured by placing a circular ROI (1.43 cm²) per phantom vial. Coefficient of variation was calculated as CV=mean/SD.
Results
Figure 1 shows the simulation results for SNR values between 20 and 2. For the investigated range of ADCnom, the fitted ADC values are plotted as mean ADCfit and SD. The simulations predict increasingly negative bias, i.e., a deviation below ADCnom, and a loss of precision, i.e., higher SD, towards higher ADCnom and towards lower SNR. ADC maps obtained in setup 1 show a progressive increase of noise in the ADC maps that is strongest in the center of the phantom (Figure 2) related to lower receiver coil sensitivity and g-factor based noise enhancement. Regarding bias and precision of the measured ADC values (Figure 3), this translates into increasingly negative bias and loss of precision from a SENSE acceleration of 4 onwards. The effect is stronger for higher ADCnom and for vials that are situated in an inner or central location of the phantom.ADC maps obtained in setup 2 show low and high noise for SENSE reconstruction at acceleration factors of 3 and 6, respectively (Figure 4). C-SENSE and C-SENSE AI recover the central ADC drop that is present for the latter case. Qualitatively, SNR is increased by C-SENSE and more so by C-SENSE AI, which makes phantom contours, but also aliasing artefacts more visible. Regarding bias and precision of the measured ADC values, bias is low for all reconstructions for an acceleration of 3 (Figure 5). At an acceleration of 6, C-SENSE and C-SENSE AI mostly recover the negative bias present in SENSE reconstruction. Moreover, these techniques lower the CV, which is brought back to a similar range as for the lower acceleration factor.
Discussion
In low-SNR EPI-DWI scans, SENSE parallel imaging reconstruction at high acceleration is prone to bias and loss of precision especially in areas with lower receiver coil sensitivity and/or high g-factor, i.e., where the coil elements have reduced overlap. C-SENSE and C-SENSE AI mitigate this problem and enable faster DWI scanning; however, radiologists may need to adapt visual inspection to the smoother appearance of ADC maps caused by reduced variation. While only relative changes between different ADC measurements can be assessed in vivo, phantoms with areas of homogeneous tissue and nominal ADC values thereof provide an excellent means of testing accuracy and precision.Conclusion
Compressed SENSE acceleration with deep-learning constrained reconstruction (C-SENSE AI) reduces ADC bias in diffusion MRI with comparison to the conventional fast imaging techniques. Improved accuracy and precision may benefit clinical examinations and lead to further acceleration in routine practice.Acknowledgements
References
1 Walker-Samuel, et al. Robust Estimation of the Apparent Diffusion Coefficient (ADC) in Heterogeneous Solid Tumors. Magn Reson Med 2009; 62:420-429.
2 Yoneyama M, et al. SNR boost in whole-body DWIBS utilizing deep learning constrained Compressed SENSE reconstruction. ISMRM 2022, p3778.
3 Yoneyama M, et al. SNR enhancement in rapid high b-value prostate single-shot DW-EPI utilizing deep learning constrained Compressed SENSE reconstruction. ISMRM 2022, p4758.
4 Meister RL, et al. Compressed SENSE in Pediatric Brain Tumor: MR Imaging Assessment of Image Quality, Examination Time and Energy Release. Clin Neuroradiol 2022; 32(3):725-733.
5 Bode M, et al. Liver diffusion‑weighted MR imaging with L1‑regularized iterative sensitivity encoding reconstruction based on single‑shot echo‑planar imaging: initial clinical experience. Sci Reports 2022;12:12468.
6 Pezzotti N, et al. An Adaptive Intelligence Algorithm for Undersampled Knee MRI Reconstruction. IEEE Access. 2020;8:204825-204838.
Figures