2202
VUDU-SAGE: Efficient T2 and T2* Mapping using Joint Reconstruction for Motion-Robust, Distortion-Free, Multi-Shot, Multi-Echo EPI1Athinoula A. Martinos Center for Biomedical Imaging, Charlestown, MA, United States, 2Harvard Medical School, Boston, MA, United States, 3Computer Engineering, Hongik University, Seoul, Korea, Republic of, 4Carleton University, Ottawa, ON, Canada, 5Fetal-Neonatal Neuroimaging & Developmental Science Center, Boston Children’s Hospital, Boston, MA, United States, 6Harvard/MIT Health Sciences and Technology, Cambridge, MA, United States
Synopsis
Keywords: Data Acquisition, Quantitative Imaging, T2 and T2* mapping
We demonstrate T2 and T2* mapping and joint reconstruction for multi-echo, multi-shot EPI data from a variable flip angle, blip-up and -down undersampling (VUDU) for spin and gradient echo (SAGE) acquisition. VUDU-SAGE employs a blip-up and -down acquisition strategy for correcting B0 distortion, and FLEET-ordering for motion-robust multi-shot EPI while maximizing signal using variable flip angles. VUDU-SAGE acquires five echoes consisting of two gradient-echo, two mixed, and one spin-echo contrasts. We jointly reconstruct all echoes and estimate T2 and T2* maps using Bloch dictionary matching. In-vivo experiment presents T2 and T2* map at 1x1x4mm3 resolution with a 9-second acquisition.Introduction
Variable flip, blip-up and -down undersampling (VUDU) enables motion-robust, distortion-free multi-shot EPI (ms-EPI)1,2. VUDU incorporates blip-up and -down acquisition (BUDA) strategy for distortion correction3,4, fast low angle excitation echo-planar technique (FLEET) for motion-robustness5 and variable flip angle (VFA) method for maximizing the signal6. BUDA employs interleaved blip-up/blip-down phase encoding and incorporates B0 forward-modeling into structured low-rank reconstruction to enable distortion-free and navigator-free msEPI. FLEET excites a specific slice successively and fully encodes the signal before proceeding to the next slice, which significantly reduces the acquisition time frame for each slice. VFA maximizes the signal of FLEET acquisition, e.g. 45° and 90° RF pulses for 2-shot EPI. Spin and gradient echo (SAGE) acquires multiple echoes for each shot and enables T2 and T2* mapping7.BUDA-SAGE successfully incorporated the BUDA strategy into the SAGE acquisition, and enabled distortion-free multi-contrast and quantitative imaging8,9. However, standard msEPI ordering requires a time gap of several seconds between multiple shots, which increases the vulnerability to motion. In this abstract, we introduce VUDU-SAGE for efficient T2 and T2* mapping using joint reconstruction of motion-robust, distortion-free, multi-echo msEPI.
Data/Code: https://anonymous.4open.science/r/vudu-sage/
Methods
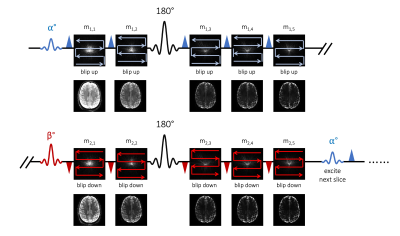
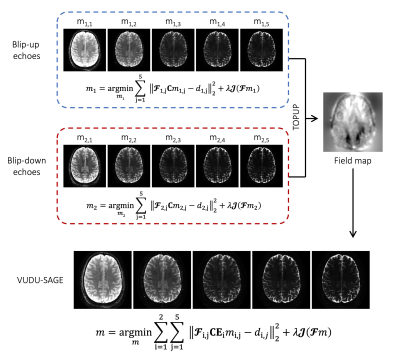
Figure 1 shows the VUDU-SAGE sequence diagram. VUDU-SAGE employs FLEET-ordering that successively excites a specific slice for motion-robust imaging5, while incorporating VFA to maximize the signal6. Each excited slice is fully encoded within a short timeframe (~320 msec) before exciting and encoding the next slice. VUDU-SAGE encodes the first-shot and the second-shot signal by a blip-up and -down readouts, respectively. VUDU-SAGE acquires five echoes consisting of two gradient-echo, two mixed, and one spin-echo contrast. Each echo covers different k-space lines to provide complementary information from each echo.Figure 2 shows the reconstruction pipeline of VUDU-SAGE. First, five echoes are jointly reconstructed for each shot. Low-rank modeling of local k-space neighborhoods (LORAKS) framework was used for the joint reconstruction as the following equation10–12.
$$m_i=\underset{m_i}{\mathrm{argmin}}\sum_{j=1}^{5}{{\left\|\mathbf{\mathcal{F}}_{i,j}\mathbf{C}m_{i,j}-d_{i,j}\right\|}_2^2}+\lambda\mathcal{J}(\mathbf{\mathcal{F}}m_i)$$
where $$$\mathbf{C}$$$ are sensitivities, $$$ \mathcal{J} $$$ is LORAKS regularization, and $$$i=1$$$ stands for blip-up and $$$i=2$$$ for blip-down. $$$\mathbf{\mathcal{F}}_{i,j}$$$, $$$m_{i,j}$$$ and $$$d_{i,j}$$$ are the undersampled Fourier transform, fully-sampled image, and acquired k-space data for $$$j^{th}$$$-echo in the the $$$i^{th}$$$ shot, respectively.
Once blip-up and -down echoes were reconstructed, we use them to estimate a field map $$$\mathbf{E}$$$, using the TOPUP algorithm13,14. Incorporating B0 forward-modeling into the joint reconstruction of two-shot, five-echo data (ten images in total), the distortion-free images can be calculated as follows.
$$m=\underset{m}{\mathrm{argmin}}\sum_{i=1}^{2}{\sum_{j=1}^{5}{{\left\|\mathbf{\mathcal{F}}_{i,j}\mathbf{C}\mathbf{E}_i m_{i,j}-d_{i,j} \right\|}_2^2}}+\lambda\mathcal{J}(\mathbf{\mathcal{F}}m)$$
On the reconstructed VUDU-SAGE images, the signal evolution obeys the following equation.
$$m(t)=\biggl\{\begin{matrix} & m^\mathrm{I}e^{-R_{2}^* t} &, 0 < t < \frac{T}{2} \\ & m^\mathrm{II} e^{-(R_{2}^*-R_{2})T}e^{-(2R_{2}-R_{2}^*)t} &, \frac{T}{2} < t < T \end{matrix} $$
where $$$T$$$ is the TE of the spin-echo contrast, $$$R_2^*={1}/{T_2^*}$$$, and $$$R_2={1}/{T_2}$$$. Voxel-wise estimates of $$$m^\mathrm{I}/m^\mathrm{II}$$$, $$$R_2^*$$$, and $$$R_2$$$ are obtained through Bloch dictionary matching on the reconstructed echoes8,9.
Experiment
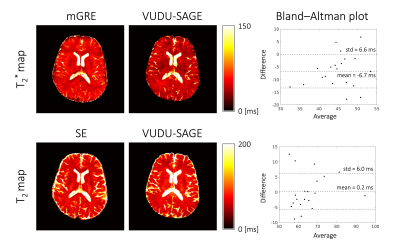
We conducted experiments on a Siemens 3T Prisma scanner equipped with a 32-channel head coil. 2-shot VUDU-SAGE acquisition was used at 8-fold acceleration per echo. The imaging parameters for VUDU-SAGE were voxel size=1x1x4mm3, partial Fourier factor = 6/8, TE= [18.11, 39.91, 87.56, 109.36, 131.16] msec, TRslice= 150 msec, slice time frame = 320 msec, and total acquisition time = 9 sec, respectively. We applied the VFA RF-pulses at flip angles $$$ \alpha $$$=37 and $$$ \beta $$$=-90, to take into account T1 recovery during the inter-shot gap6. The T2 reference map was obtained using SE acquisitions with TE=[25,50,75,100,125] msec. The T2* reference map was obtained using multi-echo gradient-recalled echo (mGRE) acquisition with TE=[4,11,18,25,32,39,46] msec. To further denoise the images, we used the image denoising network15 after the joint reconstruction. For Bland-Altman analysis, we manually selected 20 blocks consisting of 2x2 voxels.Results
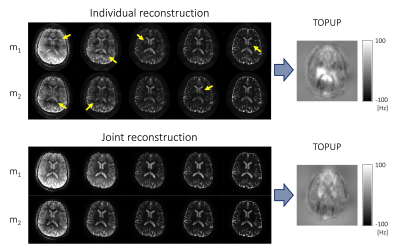
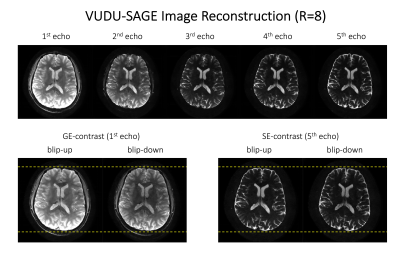
Figure 3 shows interim blip-up and -down reconstructed images for the TOPUP field-map estimation. The folding artifacts remain in the images individually reconstructed using SENSE16, due to the high acceleration, and prevent accurate field-map estimation. Joint reconstruction addressed the folding artifacts and noise amplification and robustly estimated a field map.Figure 4 shows the reconstructed distortion-free VUDU-SAGE images. The first row shows the high-fidelity five echoes. The second row demonstrates that the distortion was corrected in the reconstructed images.
Figure 5 shows the references and estimated T2 and T2* maps. In Bland-Altman analysis, the estimated T2 values were well aligned with the reference. The T2* map was slightly underestimated (6.7 msec), compared with the reference.
Discussion & Conclusion
We introduce VUDU-SAGE that allows motion-robust, distortion-free T2 and T2* mapping. In-vivo experiment presents high-fidelity multi-echo images via joint reconstruction and efficient T2 and T2* mapping using 9-second VUDU-SAGE acquisition. Although the T2* map was underestimated in the in-vivo experiment, we think including the B1 field in the signal evolution model will help improve the accuracy of T2* mapping. We think VUDU-SAGE will also allow para- and dia-magnetic susceptibility mapping8. Incorporating a neural network into joint reconstruction will further improve the distortion-free images and quantitative maps17. Employing simultaneous multislice (SMS) imaging will further accelerate the imaging speed18 and employing gSlider will boost SNR and permit high isotropic resolution4,19, thereby allowing a 20-second whole-brain quantitative MRI at the voxel size of 1x1x1mm3. We expect that VUDU-SAGE can be extended to fetal, cardiac, and abdominal imaging, where motion is unpredictable and non-rigid9.Acknowledgements
This work was supported by research grants NIH R01 EB032378, R01 EB028797, R03 EB031175, U01 EB025162, P41 EB030006, U01 EB026996, and the NVidia Corporation for computing support.References
1. Cho J, Berman AJ, Gagoski B, et al. VUDU: motion-robust, distortion-free multi-shot EPI. In: Proceedings of the 29th Scientific Meeting of ISMRM. Online Conference; p. 0002.
2. Cho J, Kim TH, Gagoski B, et al. Variable Flip, Blip-Up and -Down Undersampling (VUDU) Enables Motion-Robust, Distortion-Free Multi-Shot EPI. In: Proceedings of the 30th Scientific Meeting of ISMRM. London, UK; p. 0757.
3. Liao C, Cao X, Cho J, Zhang Z, Setsompop K, Bilgic B. Highly efficient MRI through multi-shot echo planar imaging. In: Wavelets and Sparsity XVIII. Vol. 11138. International Society for Optics and Photonics; 2019. p. 1113818. doi: 10.1117/12.2527183.
4. Liao C, Bilgic B, Tian Q, et al. Distortion-free, high-isotropic-resolution diffusion MRI with gSlider BUDA-EPI and multicoil dynamic B0 shimming. Magn. Reson. Med. 2021;86:791–803 doi: 10.1002/mrm.28748.
5. Polimeni JR, Bhat H, Witzel T, et al. Reducing sensitivity losses due to respiration and motion in accelerated echo planar imaging by reordering the autocalibration data acquisition: Reducing Losses in Accelerated EPI with FLEET-ACS. Magn. Reson. Med. 2016;75:665–679 doi: 10.1002/mrm.25628.
6. Berman AJ, Grissom WA, Witzel T, et al. Segmented spin-echo BOLD fMRI using a variable flip angle FLEET acquisition with recursive RF pulse design for high spatial resolution fMRI. In: Proceedings of 28th Annual Meeting of ISMRM. Paris, France. ; 2020. p. 5236.
7. Manhard MK, Stockmann J, Liao C, et al. A multi‐inversion multi‐echo spin and gradient echo echo planar imaging sequence with low image distortion for rapid quantitative parameter mapping and synthetic image contrasts. Magn. Reson. Med. 2021;86:866–880.
8. Zhang Z, Cho J, Wang L, et al. Blip up-down acquisition for spin- and gradient-echo imaging (BUDA-SAGE) with self-supervised denoising enables efficient T2, T2*, para- and dia-magnetic susceptibility mapping. Magn. Reson. Med. 2022;88:633–650 doi: 10.1002/mrm.29219.
9. Zhang Z, Ye H, Li M, Liu H, Bilgic B. Liver-Buda-Sage: Simultaneous Whole Liver T 2 and T* 2 Mapping in one Breath-Hold. In: 2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI). IEEE; 2022. pp. 1–5.
10. Haldar JP. Low-rank modeling of local k-space neighborhoods (LORAKS) for constrained MRI. IEEE Trans. Med. Imaging 2013;33:668–681.
11. Bilgic B, Kim TH, Liao C, et al. Improving parallel imaging by jointly reconstructing multi‐contrast data. Magn. Reson. Med. 2018;80:619–632.
12. Kim TH, Bilgic B, Polak D, Setsompop K, Haldar JP. Wave‐LORAKS: Combining wave encoding with structured low‐rank matrix modeling for more highly accelerated 3D imaging. Magn. Reson. Med. 2019;81:1620–1633.
13. Andersson JLR, Skare S, Ashburner J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. NeuroImage 2003;20:870–888 doi: 10.1016/S1053-8119(03)00336-7.
14. Smith SM, Jenkinson M, Woolrich MW, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 2004;23:S208–S219.
15. Zhang K, Zuo W, Chen Y, Meng D, Zhang L. Beyond a gaussian denoiser: Residual learning of deep cnn for image denoising. IEEE Trans. Image Process. 2017;26:3142–3155.
16. Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn. Reson. Med. Off. J. Int. Soc. Magn. Reson. Med. 1999;42:952–962.
17. Kim TH, Garg P, Haldar JP. LORAKI: Autocalibrated Recurrent Neural Networks for Autoregressive MRI Reconstruction in k-Space. ArXiv190409390 Cs Eess 2019.
18. Setsompop K, Gagoski BA, Polimeni JR, Witzel T, Wedeen VJ, Wald LL. Blipped-controlled aliasing in parallel imaging for simultaneous multislice echo planar imaging with reduced g-factor penalty. Magn. Reson. Med. 2012;67:1210–1224 doi: 10.1002/mrm.23097.
19. Setsompop K, Fan Q, Stockmann J, et al. High-resolution in vivo diffusion imaging of the human brain with generalized slice dithered enhanced resolution: Simultaneous multislice (gSlider-SMS). Magn. Reson. Med. 2018;79:141–151 doi: 10.1002/mrm.26653.
Figures