2192
Super-resolution GAN network for fast quantification of proton density mapping from highly accelerated synthetic magnetic resonance imaging1School of Biological Science and Medical Engineering, Beihang University, Beijing, China, 2Department of Radiology, Beijing Friendship Hospital, Capital Medical University, Beijing, China, 3Department of Medical Engineering, Beijing Friendship Hospital, Capital Medical University, Beijing, China, 4Philips Healthcare, Beijing, China, 5Peking university Academy for Advanced Interdisciplinary Studies, Beijing, China, 6Beijing ChuiYangLiu Hospital, Beijing, China
Synopsis
Keywords: MR Fingerprinting/Synthetic MR, Image Reconstruction
Quantitative magnetic resonance imaging (qMRI) can reflect the inherent characteristics of human tissue of relaxation time and proton density, and has important value for clinical diagnostic. However, long scan times limit the use of qMRI. We propose a method to optimize fast qMRI using a super-resolution generative adversarial network, thereby reducing scan time and obtaining accurate quantitative values. The results showed that this method was able to improve the image quality of qMRI, and the quantitative values were not significantly different from those obtained in conventional acquisitions.Introduction
Quantitative magnetic resonance imaging (qMRI) is a technique that can quantify tissue properties in the human body, such as T1 and T2 relaxation times and proton density (PD) 1. Compared to conventional weighted imaging, quantitative imaging can provide more accurate and unbiased information on tissue properties, reducing diagnostic variability due to scanner differences 2. This noninvasive measurement of tissue microstructural changes is considered crucial for clinical diagnosis and brain research 3,4. However, the long acquisition time is the main problem that limits the promotion and application of qMRI in clinical practice 5. The emergence of synthetic MRI technology based on two-dimensional (2D) multi-dynamic multi-echo (MDME) sequences can reduce acquisition time, as various images including quantitative maps can be obtained from a single acquisition 6,7. Although synthetic MRI has demonstrated better scan efficiency than conventional qMRI techniques, further acceleration is needed to translate it into routine clinical practice, especially in pediatric and poorly cooperating patient imaging.Methods
MR Datasets154 healthy volunteers were recruited and had two synthetic MRI scans using a SIGNA Pioneer 3.0T MRI scanner (GE Helthcare, Maukesha, Wis): routine scan and fast scan (low-resolution, LR). Most scan parameters remain the same between two scans: TR = 4000ms, TE1 = 18ms, TE2 = 90ms, TI = 28.2ms, thickness = 5mm, 24 slices. Matrix and acceleration factors differ between routine scan and fast scan (320 × 256 versus 192 × 128; 2 versus 3, respectively). The final scan times for routine scan and fast can were 4min55sec and 1min52sec, respectively. After MRI scanning, the raw DICOM data were loaded into Synthetic MRI software (SyntheticMR AB, version 8.0.4, Linköping, Sweden), and PD maps of two scans were retrieved.
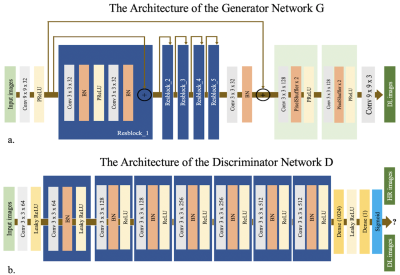
Network Architectures
The SRGAN contain a super-solution generator G and a VGG based discriminator D. The network structure is shown in Figure 1. Cross-entropy loss was used in VGG to distinguish the generated sample from ground truth. PD Maps from routine scan are taken as the ground truth high-resolution (GT) images, while PD images from fast scan are low-resolution images. The SRGAN network was trained on 139 subjects on a graphic processor (Tesla V100, NVIDIA, Santa Clara, CA, USA) for 1k epochs with a batch size of 8 using the TensorFlow framework. The parameters of the network were optimized using the Adam optimizer via stochastic gradient descent with a learning rate of 1e-3. Images from the remaining 15 subjects were used for testing. In order to demonstrate the benefit of the adversarial training, images using only the generative network (w/) are also output.
Evaluation and Statistical Analysis
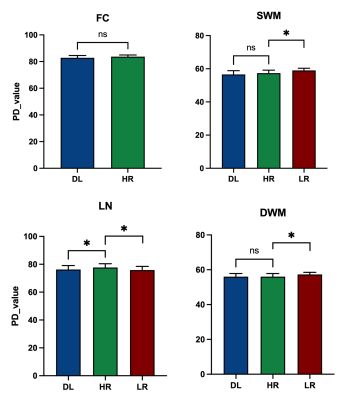
Peak Signal to Noise Ratio (PSNR) and Structural Similarity Index (SSIM) were calculated for image quality assessment. To obtain brain regional quantitative values, the PD maps were evaluated by region of interest (ROI) analysis. We used ImageJ software 8 to delineate ROIs on HR, LR and deep learning images (DL) and record the corresponding PD values. The four regions selected were frontal cortex, subcortical white matter, lenticular nucleus and deep white matter. The differences of PD values between two groups (HR vs. LR and HR vs. DL) were assessed using Wilcoxon signed rank test (paired) under GraphPad Prism (Version 9.1.1, GraphPad Software, LLC). The significance level is set to P < 0.05.
Results and Discussions
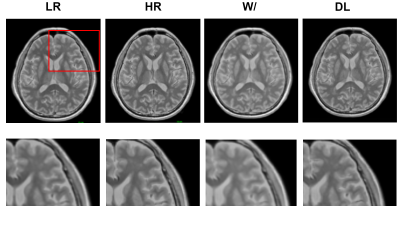
Experimental results show that the quality of PD maps is significantly improved after GAN, especially after using adversarial training (Figure 2), the images trained with GAN avoid the marginal haziness while the images trained without discriminator lost sharp details. The results of PSNR and SSIM are shown that the image quality after using the adversarial training is comparable to the HR image (58.93 ± 3.49, and 0.99 ± 0.008, respectively). Figure 3 shows the results of ROI analysis of PD values. For the frontal cortex, the LR images were too blurry to discern, and there was no significant difference between HR and DL (p = 0.15). The PD values of subcortical white matter in LR images were significantly different from HR images (p = 0.03), but there was no difference between DL images and HR images (p = 0.56). Results for deep white matter are consistent with subcortical white matter (p = 0.03 for LR images, and P = 0.89 for DL images). There are significant differences between LR/DL images and HR images in the lenticular nucleus region (p = 0.01 versus p = 0.03, respectively).Conclusions
The quality of the synthesized quantitative magnetic resonance images decreased after the scan time was shortened. By using GAN, the image quality can be effectively improved, and the quantitative value is almost indistinguishable from the conventional scan value. This study preliminarily achieves the goal of rapidly quantifying proton density from highly accelerated synthetic magnetic resonance imaging, which is helpful for the popularization and application of quantitative magnetic resonance imaging.Acknowledgements
This work was supported by Beijing Scholar 2015 (Zhenchang Wang).References
1. Shtangel, O. & Mezer, A. A. A phantom system for assessing the effects of membrane lipids on water proton relaxation. NMR Biomed. 33, 1–17 (2020).
2. Gräfe, D., Frahm, J., Merkenschlager, A., Voit, D. & Hirsch, F. W. Quantitative T1 mapping of the normal brain from early infancy to adulthood. Pediatr. Radiol. 51, 450–456 (2021).
3. Gracien, R.-M. et al. Changes and variability of proton density and T1 relaxation times in early multiple sclerosis: MRI markers of neuronal damage in the cerebral cortex. Eur. Radiol. 26, 2578–2586 (2016).
4. Vanderhasselt, T. et al. Synthetic MRI demonstrates prolonged regional relaxation times in the brain of preterm born neonates with severe postnatal morbidity. NeuroImage Clin. 29, 102544 (2021).
5. Fang, Z. et al. Deep Learning for Fast and Spatially-Constrained Tissue Quantification from Highly-Accelerated Data in Magnetic Resonance Fingerprinting. IEEE Trans. Med. Imaging 38, 2364–2374 (2019).
6. Warntjes, J. B. M., Dahlqvist, O. & Lundberg, P. Novel method for rapid, simultaneous T1, T2*, and proton density quantification. Magn. Reson. Med. 57, 528–537 (2007).
7. Warntjes, J. B. M., Dahlqvist Leinhard, O., West, J. & Lundberg, P. Rapid magnetic resonance quantification on the brain: Optimization for clinical usage. Magn. Reson. Med. 60, 320–329 (2008).
8. Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of Image Analysis. Fundam. Digit. Imaging Med. 9, 671–675 (2012).
Figures