2162
A Brain Age Estimation Network based on QSM using the Segment Transformer1School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China
Synopsis
Keywords: Quantitative Imaging, Quantitative Susceptibility mapping
Quantitative Susceptibility Mapping (QSM) reflects various biological procedures, e.g., iron accumulation in deep gray matter, which is tightly associated with brain development and highly related to various neurodegenerative diseases. Nowadays, deep-learning-based methods have achieved great success in medical image segmentation due to their ability to extract multi-scale features. We create a novel network to segment key brain areas on QSM images to improve brain age prediction. Our network further integrates multi-level features by utilizing a variant of Vision Transformer, termed Segment Transformer. Results show that our method can improve brain age estimation compared to previous studies based on T1w MRI.Introduction
Brain age is known as an informative biomarker in brain health1. The difference between chronological and brain age is highly associated with many neural system diseases, such as Parkinson's syndrome and Alzheimer's disease. Quantitative Susceptibility Mapping (QSM) can quantify the distribution of magnetic susceptibility of the biological tissues and has great potential to quantitatively investigate biological procedures of various neurodegenerative diseases2. For example, the quantity of iron accumulated in the deep gray matter can be measured by QSM and could contribute to the accurate estimation of brain age.Recently, deep learning algorithms have achieved noticeable success in plenty of medical tasks. The framework of a popular medical network, the U-Net3, has remarkable capabilities of generating concrete features and obtaining excellent segmentation performance. Those features extracted in the segmentation network are beneficial to other missions as the network extracts both low-level and high-level information.
Inspired by the strong correlation between the iron deposition in deep gray matter and brain aging and the rich contexts acquired by brain segmentation, we design a multi-task model to accomplish the missions of brain area division and brain age estimation.
Method
There are two stages of training, the segmentation pre-training, and the age estimation finetuning, shown in Figure 1.In the first stage, we used the U-Net structure to classify each voxel into nine categories, including five deep gray matter regions (globus pallidus (GP), caudate nucleus (CN), putamen (PUT), red nucleus (RN), and substantia nigra (SN)), white matter, gray matter (except the above regions), cerebrospinal fluid, and background. The encoder uses the Selective Kernel Convolution5 and channel attention and spatial attention6 for the deeper excavation of features.
In the second stage, the network connects the features generated after the bottleneck and those before the final output layer to better concatenate global information and concrete local details. We use the segmentation mask generated in the first stage to select low-level features in deep gray matter regions online and combine them with those in high-level by Segment Transformer. Segment Transformer is a variant of the Vision Transformer4 we design for better exploring latent information, which includes six layers. In each layer, the global and local features are projected into Query, Key, and Value tokens respectively (as Figure 1), and accomplish the computation of Multi-Head Cross Attention (MHCA)7 following Equation (1).
$$F^{O}_{g} = softmax(\frac{F^{Q}_{g}(F^{K}_{l})^{T}}{\sqrt{d}})F^{V}_{l}\tag{1}$$
The output tokens are concatenated with the global features, put into the MLP, and added to the global features for the final outputs used in the later layers. Bonded with three volume ratios (white matter, gray matter, and cerebrospinal fluid) and gender, the outcomes of the Segment Transformer go through the fully-connected layers to generate the brain age prediction.
The objection function is a unity of segmentation loss and age prediction loss as Equations (2)~(7),
$$Loss=λMAE+ECELD\tag{2}$$
$$MAE=\lvert \hat{y}-y\rvert\tag{3}$$
$$ECELD=(1-α)ECE-αlog(Dice)\tag{4}$$
$$Dice=\frac{2\sum_{i}^{L}\sum_{j}^{H}\sum_{k}^{W}p_{ijk}q_{ijk}}{\sum_{i}^{L}\sum_{j}^{H}\sum_{k}^{W}p_{ijk} + \sum_{i}^{L}\sum_{j}^{H}\sum_{k}^{W}q_{ijk}}\tag{5}$$
$$ECE =- \frac{\sum_{i}^{L}\sum_{j}^{H}\sum_{k}^{W}\omega_{ijk}\sum_c^{M}(q_{ijkc}(1-p_{ijkc})^{\gamma}logp_{ijkc}+(1-q_{ijkc})p_{ijkc}^{\gamma}log(1-p_{ijkc}))}{\sum_{i}^{L}\sum_{j}^{H}\sum_{k}^{W}(0.5+\omega_{ijk})}\tag{6}$$
$$\omega_{ijk}=1+0.5\underset{s\in S}{\sum}\lvert\frac{\underset{l,h,w\in A_{ijk}}{\sum}q_{ijk}^{s}}{\underset{l,h,w\in A_{ijk}}{\sum} 1}-q_{ijk}\rvert\tag{7}$$
where y, $$$\hat{y}$$$, p and q represent the chronological age, the predicted brain age, the predicted voxel label, and the true voxel label respectively. We set λ as zero during the pre-training and one during the finetuning. We use Mean Absolute Error (MAE) as the age prediction loss and Edge-weighted Cross Entropy Log Dice (ECELD)8 as the segmentation loss, which enhances the weights of edge voxels and is more friendly to small labels. We implement five-fold cross-validation and ensemble the five models by averaging their predictions on every sample.
Data acquisitions
714 healthy subjects aged from 18 to 80 years were acquired from three different scanners. The parameters of scanner 1: resolution - 0.5mm×0.5mm×2mm, TR – 33.7ms, TE1/spacing/TElast – 4.5ms/3.6ms/30.0ms. The parameters of scanner 2: resolution - 0.94mm×0.94mm×2.8mm, TR – 45ms, TE1/spacing/TElast – 5.0ms/4.9ms/39.0ms. The parameters of scanner 3: resolution - 0.65mm×0.65mm×2mm, TR – 34.6ms, TE1/spacing/TElast – 3.3ms/3.7ms/29.2ms. 165 cases are selected as the testing dataset, while the 549 samples are used in the training of five-fold cross-validation.Experiments and Results
Our model shows an average MAE of 3.9 years on the whole testing dataset for brain age prediction (Table 1). We also used T1-weighted MRIs and registered QSMs as inputs to compare the effects of different modalities. Our experiments prove that every strategy, including segmentation pre-training, Segment Transformer, DGM cropping, and QSM of individual space is inevitable.Discussion and Conclusion
As a novel kind of quantitative imaging method, QSM images contain more concrete and abundant details that are conducive to medical judgments. Meanwhile, deep learning methods can explore information that is hard to represent in traditional approaches. With the prior knowledge in previous research, our network achieves online brain area segmentation to achieve an improved brain age estimation. The contribution heatmaps in Figure 3 indicate that our model mainly focuses on reasonable brain regions2 to generate the final prediction. Our model pays more attention to five DGM areas for young people (ages between 18 and 45), but concentrates on SN, RN, and some parts of the cortex for old people (ages between 46 and 80). Using images in the individual space not only accelerates the computation but also avoids potential errors involved in registration. Both QSM and Deep learning are emerging technologies for clinical diagnoses and treatments, and we merge them successfully in a complementary way.Acknowledgements
References
1. J. H. Cole and K. Franke, Predicting age using neuroimaging: Innovative brain ageing biomarkers. Trends Neurosci., vol. 40, no. 12, pp. 681–690, Dec. 2017.
2. Liu, Chunlei et al. Quantitative Susceptibility Mapping: Contrast Mechanisms and Clinical Applications. Tomography 1 (2015): 3 - 17.
3. Ronneberger, Olaf et al. U-Net: Convolutional Networks for Biomedical Image Segmentation. ArXiv abs/1505.04597 (2015): n. pag.
4. Li, Xiang et al. Selective Kernel Networks. 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) (2019): 510-519.
5. Woo, Sanghyun et al. CBAM: Convolutional Block Attention Module. ECCV (2018).
6. Dosovitskiy, Alexey et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. ArXiv abs/2010.11929 (2021): n. pag.
7. Wen, Zhengyao et al. Distract Your Attention: Multi-head Cross Attention Network for Facial Expression Recognition. ArXiv abs/2109.07270 (2021): n. pag.
8. Lee, Min Seok et al. TRACER: Extreme Attention Guided Salient Object Tracing Network. AAAI (2022).
Figures
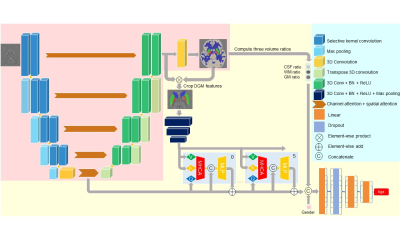
Figure 1. The structure of our network. In the first stage, we only pre-train blocks in the pink background to generate segmentation maps for input images. During the fine-tuning, we train the whole network together to guarantee precise outputs of age prediction and segmentation mask. We detach the gradient backpropagation before cropping DGM features to keep the segmentation map accurate and stable in the second stage. For simplicity, we only draw two layers of the Segment Transformer, but use six layers in total.
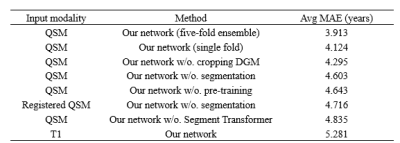
Table 1. The experiment results of our study. The proposed model was obtained by five-fold cross-validation and all ablation studies are based on the same fold for convenience.
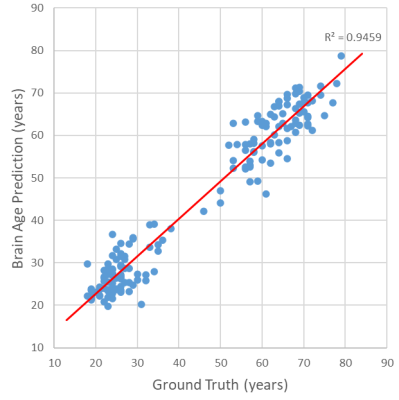
Figure 2. The scatter plot of our ensembled model’s output. Every blue point represents a sample in the testing dataset. The red line is the linear regression curve of the predicted brain age to the corresponding ground truth, and its R2 is 0.95.
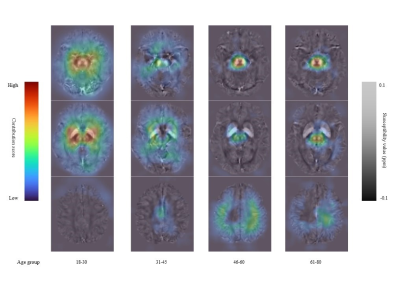
Figure 3. The contribution heatmaps of our model. We divide the whole testing dataset into four groups according to their ages, considering the divergence of brain conditions of different ages. The pictures above show average results in each group. Susceptibility value contributing more to the brain age is colored coded by dark red.