2155
SSL-QALAS: Self-Supervised Learning for Multiparametric Quantitative MRI Using QALAS1Athinoula A. Martinos Center for Biomedical Imaging, Charlestown, MA, United States, 2Department of Radiology, Harvard Medical School, Boston, MA, United States, 3Department of Radiology, Massachusetts General Hospital, Boston, MA, United States, 4Fetal-Neonatal Neuroimaging & Developmental Science Center, Boston Children’s Hospital, Boston, MA, United States, 5Harvard/MIT Health Sciences and Technology, Cambridge, MA, United States
Synopsis
Keywords: Quantitative Imaging, Quantitative Imaging
The 3D-quantification using an interleaved Look-Locker acquisition sequence with T2 preparation pulse (3D-QALAS) has been developed and used for acquiring high-resolution T1, T2, and PD maps from five measurements. The dictionary matching method can be used for generating quantitative maps from the acquired multi-contrast images; however, it requires an external dictionary, which needs to be pre-calculated and voxel-by-voxel fitting is computationally demanding. In this study, we propose to generate multiple quantitative maps including T1, T2, PD, and inversion efficiency (IE) maps using self-supervised learning from 3D-QALAS measurements (i.e., SSL-QALAS) for rapid, accurate, and dictionary-free multiparametric fitting.Introduction
Quantitative MRI (qMRI) provides quantitative tissue information including T1, T2, T2* relaxation rates, and proton density (PD), which can be used for tissue analysis and diagnosis of abnormalities such as brain tumors and multiple sclerosis1-3. The 3D-quantification using an interleaved Look-Locker acquisition sequence with T2 preparation pulse (3D-QALAS) has been developed and used for acquiring high-resolution T1, T2, and PD maps from five measurements within each repetition time (TR)4-6. Bloch-simulation based dictionary matching can be used for generating quantitative maps from the acquired multi-contrast images; however, it requires an external dictionary, which needs to be pre-calculated and requires a long computation time for voxel-by-voxel fitting. Recently, self-supervised learning (SSL) that does not require external training data has been utilized for MRI denoising, reconstruction, and quantitative mapping7-9. In this study, we propose to generate multiple quantitative maps including T1, T2, PD, and inversion efficiency (IE) maps using self-supervised learning from 3D-QALAS measurements (i.e., SSL-QALAS) for rapid and dictionary-free multiparametric fitting.Example Code: https://anonymous.4open.science/r/SSL-QALAS/
Methods & Experiments
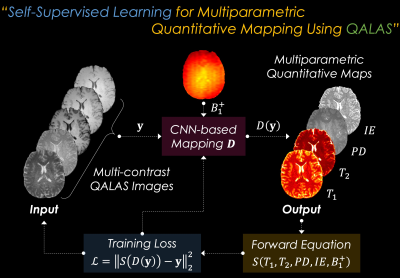
Self-Supervised Learning for Multiparametric Fitting: The overall flowchart of the proposed SSL scheme for multiparametric quantitative mapping is presented in Fig. 1. The SSL-QALAS mapping is based on convolutional neural network (CNN) architecture. The five acquired QALAS contrast images ($$$y$$$) along with a separately acquired B1+ map are fed into the model ($$$D$$$) as the input. This model then estimates T1, T2, PD, and IE maps as the output ($$$D(y)$$$). The SSL scheme is enabled by calculating the l2 loss $$$L$$$ between the acquired images and synthetic images which are generated by feeding the output maps into the forward Bloch equation $$$S$$$:$$L=\left\|S(D(y))-y\right\|^2_2,$$
where $$$S$$$ is the QALAS Bloch equation, which is dependent on T1, T2, PD, IE, and B1+ maps.
Deep Learning Architecture: The proposed architecture consists of 5 CNN layers where each layer has 1x1 convolutional layer and ReLU activation function. The last layer has Softmax activation function with constant multiplication for generating 4 different outputs including T1, T2, PD, and IE, which helps the output to be in user-specified physical ranges. The model was trained with Adam optimizer. The deep learning model including QALAS Bloch equations was implemented using PyTorch library.
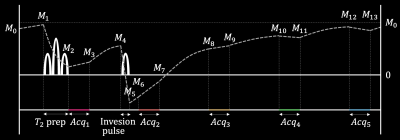
Dictionary Matching: The dictionary was generated based on QALAS Bloch equations4 with the following T1, T2, and IE ranges: T1=[5–5000ms], T2=[1–2500ms], and IE=[0.5–1.0]. For the dictionary matching, it took up to 5 minutes for each slice using 11 CPU workers. The sequence diagram of QALAS is presented in Fig. 2.
Data Acquisition: We acquired in vivo data from a healthy volunteer using 3D-QALAS sequence from a 3T MAGNETOM Prisma scanner (Siemens Healthcare, Erlangen, Germany) with a 32ch head receive array. The detailed scan parameters for the 3D-QALAS are as follows: FOV=228x228x208mm3, matrix size=176x176x160, BW=320Hz/pixel, echo spacing=5.7ms, turbo factor=128, TR=4.5s, TE=2.3ms, acceleration R=2, and scan time=6m 34s. We scanned the same volunteer using 3D-QALAS sequence for 4 times: initial scan, re-scan (without any changes), re-landmark (altering the landmark position), and re-position (repositioning the volunteer). B1+ maps were additionally acquired using a magnetization-prepared turbo-fast low angle shot (turbo-FLASH) sequence10 using the following scan parameters: FOV=228x228x208mm3, matrix size=64x64x58, BW=490Hz/pixel, TR=12.68s, TE=2.66ms, acceleration R=2, and scan time=27s.
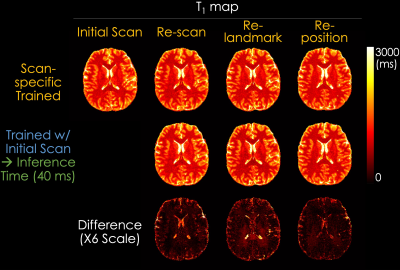
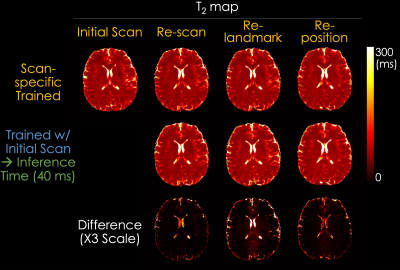
Experiments: We evaluated the dictionary matching model and our proposed SSL method by comparing the estimated T1, T2, PD, and IE maps. In addition, to validate the generalizability of the SSL method, we compared the maps reconstructed using a scan-specific trained model and a pre-trained model with initial data. In the scan-specific model, the training and inference data are identical. The pre-trained model was trained with initial data and rapidly inferred (~40 ms for inferring each slice using a single RTX 5000 GPU) to other acquisitions including re-scan, re-landmark, and re-position.
Results
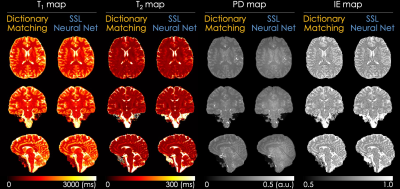
Fig. 3 shows the T1, T2, PD, and IE maps reconstructed using the dictionary-based matching and the proposed SSL method. The SSL method generates quantitative maps comparable with those reconstructed from a dictionary-based method, which indicates that the quantitative maps can be obtained using a neural network and QALAS forward equations without an external dictionary. Fig. 4 and Fig. 5 present the reconstructed T1 and T2 maps using the proposed SSL method with 4 different acquisitions (i.e., initial, re-scan, re-landmark, and re-position) from the same subject. The maps in the first row were reconstructed using a scan-specific trained model (i.e. network trained from scratch for each dataset) while those in the second row were reconstructed using the model that was trained with initial scan data. The difference images show that the T1 and T2 maps of the scan-specific model and pre-trained model are similar except for CSF regions. For T2 maps, the errors in CSF are higher than in other regions such as white and gray matter.Discussion & Conclusion
In this study, we demonstrated that rapid and dictionary-free multiparametric quantitative MRI including T1, T2, PD, and IE maps can be obtained using the SSL scheme from 3D-QALAS measurements. The proposed scheme is not only able to be applied to a scan-specific strategy but also able to be extended to a general model that can be used for other scan data by fine-tuning the pre-trained model instead of training the model from scratch.Acknowledgements
This work was supported by research grants NIH R01 EB032378, R01 EB028797, R03 EB031175, U01 EB025162, P41 EB030006, U01 EB026996, and the NVidia Corporation for computing support.References
[1] Keenan, K.E., Biller, J.R., Delfino, J.G., Boss, M.A., Does, M.D., Evelhoch, J.L., Griswold, M.A., Gunter, J.L., Hinks, R.S., Hoffman, S.W. and Kim, G., 2019. Recommendations towards standards for quantitative MRI (qMRI) and outstanding needs. Journal of magnetic resonance imaging: JMRI, 49(7), p.e26.
[2] Granziera, C., Wuerfel, J., Barkhof, F., Calabrese, M., De Stefano, N., Enzinger, C., Evangelou, N., Filippi, M., Geurts, J.J., Reich, D.S. and Rocca, M.A., 2021. Quantitative magnetic resonance imaging towards clinical application in multiple sclerosis. Brain, 144(5), pp.1296-1311.
[3] Tofts, P. ed., 2005. Quantitative MRI of the brain: measuring changes caused by disease. John Wiley & Sons.
[4] Kvernby, S., Warntjes, M.J.B., Haraldsson, H., Carlhäll, C.J., Engvall, J. and Ebbers, T., 2014. Simultaneous three-dimensional myocardial T1 and T2 mapping in one breath hold with 3D-QALAS. Journal of Cardiovascular Magnetic Resonance, 16(1), pp.1-14.
[5] Fujita, S., Hagiwara, A., Hori, M., Warntjes, M., Kamagata, K., Fukunaga, I., Andica, C., Maekawa, T., Irie, R., Takemura, M.Y. and Kumamaru, K.K., 2019. Three-dimensional high-resolution simultaneous quantitative mapping of the whole brain with 3D-QALAS: an accuracy and repeatability study. Magnetic resonance imaging, 63, pp.235-243.
[6] Fujita, S., Hagiwara, A., Hori, M., Warntjes, M., Kamagata, K., Fukunaga, I., Goto, M., Takuya, H., Takasu, K., Andica, C. and Maekawa, T., 2019. 3D quantitative synthetic MRI‐derived cortical thickness and subcortical brain volumes: Scan–rescan repeatability and comparison with conventional T1‐weighted images. Journal of Magnetic Resonance Imaging, 50(6), pp.1834-1842.
[7] Tian, Q., Li, Z., Fan, Q., Polimeni, J.R., Bilgic, B., Salat, D.H. and Huang, S.Y., 2022. SDnDTI: Self-supervised deep learning-based denoising for diffusion tensor MRI. NeuroImage, 253, p.119033.
[8] Akçakaya, M., Yaman, B., Chung, H. and Ye, J.C., 2022. Unsupervised Deep Learning Methods for Biological Image Reconstruction and Enhancement: An overview from a signal processing perspective. IEEE Signal Processing Magazine, 39(2), pp.28-44.
[9] Liu, F., Kijowski, R., El Fakhri, G. and Feng, L., 2021. Magnetic resonance parameter mapping using model‐guided self‐supervised deep learning. Magnetic resonance in medicine, 85(6), pp.3211-3226.
[10] Chung, S., Kim, D., Breton, E. and Axel, L., 2010. Rapid B1+ mapping using a preconditioning RF pulse with TurboFLASH readout. Magnetic resonance in medicine, 64(2), pp.439-446.
Figures