2130
Attention Deep-Shallow Network (ADSN): A Deep Learning Model for IDH and TERTp Mutation Detection in Gliomas using 1H-MRS1Institute of Biomedical Engineering, Bogazici University, Istanbul, Turkey, 2Brain Tumor Research Group, Acibadem University, Istanbul, Turkey, 3Department of Medical Pathology, Acibadem University, Istanbul, Turkey, 4Department of Biomedical Engineering, Acibadem University, Istanbul, Turkey, 5Department of Neurosurgery, Acibadem University, Istanbul, Turkey, 6Department of Radiology, Acıbadem University, Istanbul, Turkey
Synopsis
Keywords: Tumors, Machine Learning/Artificial Intelligence, Deep Learning
Isocitrate dehydrogenase (IDH) and telomerase reverse transcriptase promoter (TERTp) mutations affect the clinical behavior and survival rate of diffuse gliomas. According to the latest WHO 2021 brain tumor classification, IDH mutation is an important factor for grouping adult-type diffuse gliomas. The preoperative detection of these mutations is very critical for treatment planning. In this study, we propose enhanced 1D-CNN models by adding an attention mechanism as a prior network to focus on relevant spectral frequencies of 1H-MRS to identify IDH-mutant (IDH-mut), TERTp-mutant (TERTp-mut), and IDH-wt, TERTp-mut (TERTp-only) gliomas using three binary modelsSummary of Main Findings
An attention mechanism focusing on relevant spectral frequencies of 1H-MRS enhanced the performance of 1D-CNN models resulting in F1 scores of 93% for IDH mutant, 90% for TERTp mutant, and 80% for IDH wildtype, TERTp mutant glioma identification.Introduction
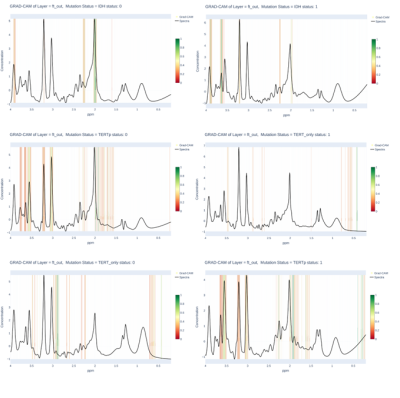
Molecular genetic features become dramatically important for understanding the underlying biology of brain tumors. World Health Organization (WHO) has included genetic features in the brain tumor classification guidelines starting from 2016 [1, 2]. IDH and TERTp mutations affect survival rates and prognosis of gliomas [3, 4]. Proton magnetic resonance spectroscopy (1H-MRS) has been used for preoperative and noninvasive detection of these mutations [5-8]. The aim of this study is to employ a deep learning approach to identify IDH and TERTp mutations in gliomas based on 1H-MRS. We propose enhanced 1D-CNN models by adding an attention mechanism as a prior network to identify relevant spectral frequencies of the 1H-MRS before classification. Additionally, gradient-weighted class activation mapping (Grad-CAM) [9] was used as an Explainable Artificial Intelligence (XAI) method to get insights into our proposed models' decision making processes.Methods
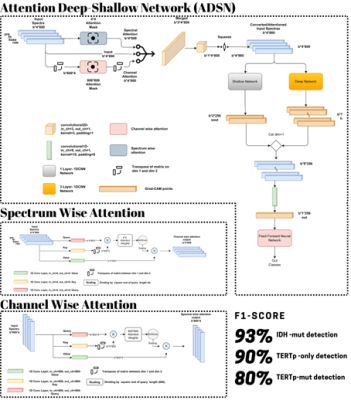
A total of 207 patients diagnosed with diffuse gliomas (mean age: 45±14.21 years, median: 43 years, range: 20-84 years, 131 males/76 females) were included in this IRB approved study. The patients were scanned before surgery at a 3T Siemens scanner (Erlangen, Germany) using a 32-channel head coil. MRS data were acquired from the solid tumor region excluding necrosis, edema, and hemorrhage using a Point Resolved Spectroscopy (PRESS) sequence (TR/TE=2000/30 ms, voxel size=1-8 cm3). We used Optuna for hyperparameter optimization [10]. There are four main parts in our pipeline, which were preprocessing, attention mechanism, deep line, and shallow line. First of all, we preprocessed data by using smoothing -Savitzky-Golay filter ws.11-, power transformation -Yeo-Johnson- and standard scaling. Two different attention mechanisms were used. Using multiple channels may cause an increase in not only relevant but also non-relevant data. As a result of that, we used not only spectrum attention but also channel attention to get the most relevant data in both axes to increase the performance of the 1D-CNN model. The third component was the deep line, which was basically a 1D-CNN network with three convolutional layers to catch finer details. The last component was the shallow line, which was also a 1D-CNN network with one layer to catch high-level features.Results
The results of the performance evaluation metrics are shown in Table 1. The grad-CAM output of attention layer in Figure 1 shows the spectral frequencies that were paid attention by each model for different glioma mutational subgroup classifications. Our models achieved 93% F1-score for identifying IDH mutation, 80% F1-Score for predicting TERTp mutation, and 90% F1-score for identifying IDH-wt, TERTp-mut gliomas on the test sets.Discussion and Conclusion
Previous studies indicated that IDH mutation could be detected with 94% accuracy with a deep learning model without employing attention layers, while TERTp mutation detection accuracy was only 76% [11]. In this study, adding attention layers to the deep learning networks especially increased the accuracy of TERTp mutation detection in gliomas. The usage of attention layers is quite new for deep learning models that are used to identify genetic mutations based on 1H-MRS or other MRI modalities, and adapting such successful solutions is valuable. Moreover, supporting the attention network using XAI -Grad-CAM-, shows the relevant peaks and/or regions of the spectra, which might lead researchers to discover possible new radiological features of genetic mutations in gliomas.Acknowledgements
This study was supported by TUBITAK 1003 grant 216S432.References
1. Louis, D.N., et al., The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathologica, 2016. 131(6): p. 803-820.
2. Louis, D.N., et al., The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol, 2021. 23(8): p. 1231-1251.
3. Eckel-Passow, J.E., et al., Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N Engl J Med, 2015. 372(26): p. 2499-508.
4. Akyerli, C.B., et al., Use of telomerase promoter mutations to mark specific molecular subsets with reciprocal clinical behavior in IDH mutant and IDH wild-type diffuse gliomas. J Neurosurg, 2018. 128(4): p. 1102-1114.
5. Choi, C., et al., 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med, 2012. 18(4): p. 624-9.
6. Nagashima, H., et al., Diagnostic value of glutamate with 2-hydroxyglutarate in magnetic resonance spectroscopy for IDH1 mutant glioma. Neuro-Oncology, 2016. 18(11): p. 1559-1568.
7. Branzoli, F., et al., Highly specific determination of IDH status using edited in vivo magnetic resonance spectroscopy. Neuro-Oncology, 2018. 20(7): p. 907-916.
8. Ozturk-Isik, E., et al., Identification of IDH and TERTp mutation status using (1) H-MRS in 112 hemispheric diffuse gliomas. J Magn Reson Imaging, 2020. 51(6): p. 1799-1809.
9. Selvaraju, R.R., et al. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. in Conference Proceedings of 2017 IEEE International Conference on Computer Vision (ICCV). 2017. p. 618-626.
10. Akiba, T., et al., Optuna: A Next-generation Hyperparameter Optimization Framework. 2019.
11. Bas A, S.-B.B., Danyeli AE, Yakicier C, Pamir MN, Ozduman K, Dincer A, Ozturk-Isik E., 1D-CNN for the Detection of IDH and TERTp Mutations in Diffuse Gliomas using Proton Magnetic Resonance Spectroscopy. International Society for Magnetic Resonance in Medicine, 2021: p. 957.