2124
Boosting The Deep Learning Performance in Predicting IDH Mutation in Gliomas Using Multiparametric MRI Including SWI, FLAIR and CE-T1WI1Institute of Biomedical Engineering, Bogazici University, Istanbul, Turkey, 2Basaksehir Cam and Sakura City Hospital, Istanbul, Turkey, 3Electric and Electronic Engineering Department, Bogazici University, Istanbul, Turkey, 4Brain Tumor Research Group, Acibadem University, Istanbul, Turkey, 5Center for Neuroradiological Applications and Reseach, Acibadem University, Istanbul, Turkey, 6Department of Medical Pathology, Acibadem University, Istanbul, Turkey, 7Department of Neurosurgery, Acibadem University, Istanbul, Turkey, 8Department of Radiology, Acibadem University, Istanbul, Turkey
Synopsis
Keywords: Tumors, Machine Learning/Artificial Intelligence
Gliomas with IDH mutations tend to have a better prognosis regardless of the histopathological grade. The main aim of this study was to identify isocitrate dehydrogenase (IDH) mutations in glioma patients using deep learning models based on SWI, FLAIR and CE-T1W images separately and together. As a result, a 2D CNN model based on multiparametric MRI resulted in an accuracy of 92.8%, while CNN based on just SWI had 72.2% and CNN based on just FLAIR had 61.1% accuracies for predicting IDH mutation in gliomas.Introduction
Isocitrate dehydrogenase mutation status has been reported to be an important factor in the tumor behavior of gliomas resulting in a better prognosis (1). In contrast, low-grade gliomas with wild-type IDH have been reported to have a similar prognosis to high-grade gliomas and have been recently grouped as ‘Glioblastoma IDH-wildtype’ according to the new WHO 2021 brain tumor classification guidelines (2-5). Furthermore, the prognosis of lower-grade gliomas after gross tumor resection was linked to their IDH mutation status (6). Gliomas with IDH mutation are also managed and treated differently than IDH wild-type gliomas (7). Today, the gold standard for identifying IDH mutations is to apply immunohistochemistry (IHC) or gene sequencing on a tissue specimen obtained via biopsy or surgical resection. However, invasive procedures harbor potential severe complications and incorrect results due to the heterogeneity of tumors and there is a growing interest in trying to differentiate IDH mutation status noninvasively prior to surgery. Different MRI techniques, such as conventional MRI, MR spectroscopy, and susceptibility-weighted MRI (SWI), have been reported to be beneficial in predicting IDH mutations (8-10). However, there has yet to be an agreement on which modality helps predicting IDH mutations better. Advances in deep learning can help us take advantage of all MRI modalities simultaneously, revealing features that are not visible to the naked eye and enabling us to detect the IDH mutation. This study aims to predict IDH mutation status in glioma patients by combining the findings of SWI and conventional MRI using deep learning models.Material-Methods
One hundred twenty seven patients with glioma (76M/51F), who underwent a preoperative MRI and provided written informed consent, were retrospectively recruited in this IRB approved study. Tumor samples were embedded in formalin-fixed paraffin to extract DNA followed by sequencing. MRI protocol consisted of FLAIR (TR/TE=8320/92ms, TI =2000ms, field of view (FOV) =220mm, slice thickness=3mm), SWI (TR/TE=28/20ms, FOV=220mm, slice thickness=1.6mm), and contrast-enhanced T1W MRI (CE-T1W) (TR/TE=589/10ms, FOV=220mm, slice thickness=3mm) on a 3T clinical MR scanner (Siemens Healthcare, Erlangen, Germany). The tumor volumes on FLAIR were segmented using 3D Slicer version 4.8.1 (http://slicer.org/). The high resolution SWI was used to register the CE-T1W and FLAIR images. The segmentation masks for each subject were then registered onto the SWI using the transformation matrix (FSL; http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/). The longest segmentation in both x and y directions was used to choose a rectangular bounding box around the lesion and slice on each modality. Rectangular largest lesion area on CE-T1W, FLAIR and SWI were merged to create RGB images using MATLAB (MathWorks Inc., Natick, MA, USA). Afterwards, the slice including largest lesion area, previous and next slides of selected slice on each modality were merged to create 2D RGB images for comparison of multimodality and one modality imaging. 2D RGB image inputs were resized to 224x224x3. Standard data augmentation with rotation of up to 90 and standard scaler standardization were utilized for preprocessing. Hyperparameter optimization framework was used to choose optimal architecture for this dataset (11). The model consisted of a 4-block 2D CNN. IDH mutation status was obtained from the last dense layer of sigmoid activation function. Binary cross-entropy as a loss function and accuracy were used in the validation cohort to evaluate the model performance.Results
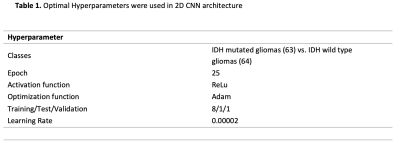
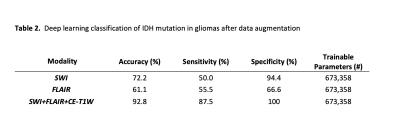
There were 64 IDH wild-type gliomas and 63 IDH mutant gliomas. The mean age of IDH mutant and wild type were 38±10, 53±13 years old, respectively. The number of patients with high (Grades 3 and 4) and low (Grades 1 and 2) grades was distributed equally in the IDH wild and mutant groups (P>0.05). Optimal hyperparameters are summarized in Table 1. In the validation set, 2D CNN based on FLAIR imaging demonstrated an accuracy of 61.1 % with a sensitivity of 55.5 % and specificity of 66.6 % in predicting IDH mutation status, while 2D CNN based on SWI imaging predicted IDH mutation status with an accuracy of 75.5%, a sensitivity of 78.2%, and a specificity of 72.7%. In contrast, 2D CNN based on multiparametric imaging demonstrated an accuracy of 92.8%, with a sensitivity of 87.5% and a specificity of 100% in predicting IDH mutation status (Table 2).Discussion-Conclusion
Our study demonstrated that conventional imaging with additional SWI might be helpful for the prediction of IDH mutation in gliomas using deep learning methods. IDH gene mutation affects glioma prognosis and treatment response irrespective of histologic grade (2). SWI is extremely sensitive to detecting blood products and calcification and can reveal neovascularization and calcification. Deep learning has shown promising results for identifying IDH mutation status using MRI, but there currently needs to be more models that incorporate multiparametric MRI at the same time (12-14). The simultaneous analysis of the SWI with conventional MR sequences (CE- T1W and FLAIR) detected IDH mutation with 92.8% accuracy. The proposed model using multiparametric MRI could boost the accuracy compared to models using only SWI and FLAIR.Acknowledgements
This study was supported by TUBITAK 1003 grant 216S432.References
1. Riemenschneider MJ, Jeuken JW, Wesseling P, Reifenberger G. Molecular diagnostics of gliomas: state of the art. Acta Neuropathol. 2010;120(5):567-84.
2. Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765-73.
3. Jiao Y, Killela PJ, Reitman ZJ, Rasheed AB, Heaphy CM, de Wilde RF, et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3(7):709-22.
4. Ostrom QT, Gittleman H, Fulop J, Liu M, Blanda R, Kromer C, et al. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2008-2012. Neuro-Oncology. 2015;17(suppl_4):iv1-iv62.
5. Nobusawa S, Watanabe T, Kleihues P, Ohgaki H. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res. 2009;15(19):6002-7.
6. Patel SH, Bansal AG, Young EB, Batchala PP, Patrie JT, Lopes MB, et al. Extent of Surgical Resection in Lower-Grade Gliomas: Differential Impact Based on Molecular Subtype. AJNR Am J Neuroradiol. 2019;40(7):1149-55.
7. SongTao Q, Lei Y, Si G, YanQing D, HuiXia H, XueLin Z, et al. IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Sci. 2012;103(2):269-73.
8. Pope WB, Prins RM, Albert Thomas M, Nagarajan R, Yen KE, Bittinger MA, et al. Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J Neurooncol. 2012;107(1):197-205.
9. Patel SH, Poisson LM, Brat DJ, Zhou Y, Cooper L, Snuderl M, et al. T2-FLAIR Mismatch, an Imaging Biomarker for IDH and 1p/19q Status in Lower-grade Gliomas: A TCGA/TCIA Project. Clin Cancer Res. 2017;23(20):6078-85.
10. Kong LW, Chen J, Zhao H, Yao K, Fang SY, Wang Z, et al. Intratumoral Susceptibility Signals Reflect Biomarker Status in Gliomas. Sci Rep. 2019;9(1):17080.
11. O'Malley TaB, Elie and Long, James and Chollet, Fran\c{c}ois and Jin, Haifeng and Invernizzi, Luca and others. KerasTuner 2019 [Available from: https://github.com/keras-team/keras-tuner.
12. Bangalore Yogananda CG, Shah BR, Vejdani-Jahromi M, Nalawade SS, Murugesan GK, Yu FF, et al. A novel fully automated MRI-based deep-learning method for classification of IDH mutation status in brain gliomas. Neuro Oncol. 2020;22(3):402-11.
13. Decuyper M, Bonte S, Deblaere K, Van Holen R. Automated MRI based pipeline for segmentation and prediction of grade, IDH mutation and 1p19q co-deletion in glioma. Comput Med Imaging Graph. 2021;88:101831.
14. Pasquini L, Napolitano A, Tagliente E, Dellepiane F, Lucignani M, Vidiri A, et al. Deep Learning Can Differentiate IDH-Mutant from IDH-Wild GBM. J Pers Med. 2021;11(4).
Figures