2123
Synthesize conventional MRI Sequences by Generative Adversarial Networks with only T2 for Use in a Multisequence gliomas classification Model1Lanzhou University Second Hospital, Lanzhou, China, 2Philips Healthcare, Xi'an, China
Synopsis
Keywords: Tumors, Machine Learning/Artificial Intelligence
The aim of this study was to test deep learning classification models of glioma subtypes using the generated images. GANs were created based on the two frameworks, pix2pix and cycleGAN. The source domain was T2 and the target domain was T1c, T2-FLAIR or ADC. The results demonstrated that the T2 to T1c pix2pix model has the highest PSNR and SSI. When only the T2-flair or T1c sequence is replaced with the generated image, the classification accuracy is same as the original image. Therefore, depending solely on T2 sequences, GANs networks could generate other sequences for Use in gliomas classification Model.Introduction
Deep learning (DL) methods have been demonstrated to noninvasively predict glioma subtypes. However, there is a strong requirement for the integrity of patient image sequences since DL classification models depend on multimodal image input. Patient images are missing in clinical work due to patient limitations, delayed image saving and uploading, and image artifacts, etc. Therefore, an image processing is required to deal with missing sequences. Generative adversarial networks (GANs) are new type of DL model that can create medical pictures. Many previous studies[1,2] used paired data (T1WI and T2WI) to generate T1c and T2-Flair by pix2pix framework (the first image-to-image GANs). Hovever, complete paired data are relatively difficult to get. Compared to pix2pix, cycleGAN is an image-to-image translation framework that does not require aligned training pairs. In order to translate images between the target domain and the source domain, cycleGAN stitches two generators together from top to bottom. CycleGAN has been shown in several studies to be an effective supplementary tool in medical pictures. This study compared the quality of medical images generated by pix2pix and cycleGAN with the same dataset size. Then, using only T2 images, we attempted to generate all conventional sequences that used to test DL classification models of glioma subtypes.Methods
Two hundred and eleven patients with gliomas were recruited from Lanzhou university second Hospital. All patients were scanned using a 3.0T MR (Ingenia CX, Philips Healthcare) with 16-channel head coil. The imaging protocol included T1-weighted post-contrast (T1c), T2-weighted (T2), T2 Fluid Attenuated Inversion Recovery (T2-FLAIR) and Diffusion weighted imaging (DWI). DWI derived Apparent Diffusion Coefficient (ADC) map was automatically generated by MR host. The middle 5-15 layers are picked from each MRI sequence of the patients for subsequent use. After excluding images with inferior image quality, a total of 2200 images were obtained and randomly divided into 1800 for training, 200 for validation, and 200 for testing. All images were resized to 256×256 dimensions, and MRI intensities were normalized to (–1, 1) range. To construct the missing MRI sequences, we created GAN models based on the two frameworks, pix2pix and cycleGAN. While cycleGAN took unpaired images as input, pix2pix took paired images instead. For all the models, the source domain was T2 and the target domain was T1c, T2-FLAIR or ADC. Figure 1 showed the training process of GANs. Peak Signal-to-Noise Ratio (PSNR) and the structural similarity index (SSI) were used to evaluate the differences of MRI signal intensities between the original images and the GAN-generated images. In addition, after replacing the original image with the generated image and feeding the data into a DL model[3] for classifying gliomas (Combinations of 3 image modalities to produce a multi-contrast RGB image), we compared the accuracy of the models with different image combination (Figure 2).Results
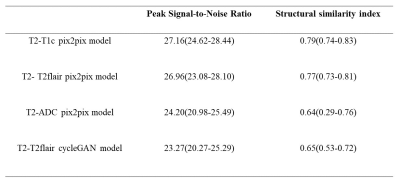
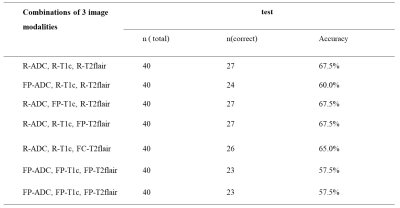
Table 1 showed the median and 25th-75th percentiles of the PSNR and the SSI for all test sets. The T2 to T1c pix2pix model has the highest PSNR and SSI (median PSNR = 27.16 and median SSI = 0.79) (Tabel 1). When only the T2-flair or T1c sequence is replaced with the generated image, the classification accuracy is the same as the model of the original image (accuracy = 67.5%). The classification accuracy reduced by 10% when only generated images were used (Table 2, Figure 3).Discussion
By depending solely on T2 sequences, GANs networks could generate other conventional sequences (T1c, T2-FLAIR or ADC). When used as input to the classification model, the generated image could achieve the same accuracy as the original image. Previous research typically used similar acquisition sequences’ parameters for paired input, such as T1 and T1c, T2 and T2-flair, to increase the quality of the generated images. Different source domains, however, required additional processing times and stricter data integrity requirements. In fact, with limited datasets, standard sequences with superior quality could be generated by using just T2 sequence (median PSNR ranged from 24.20 to 27.16 and median SSI ranged from 0.64 to 0.79). It is worth noting that the generated ADC image's quality is unstable, and its deviation is significant. One explanation is that the ADC image is derived from the DWI image, which has low resolution and a significant overall gap with the T2 sequence. To improve the gentrated ADC image’s quality, a more training data might be required. In our results, pix2pix outperformed cycleGAN on the same size of dataset. CycleGAN provided the benefit of not requiring paired data, which minimizes the difficulty of producing datasets when compared to pix2pix[4]. Nevertheless, pix2pix's training time was just about one tenth compared to cycleGAN. The two had distinct application conditions and cannot be substituted for one another. It performed approximatively in the classification model when there was still a little gap between the generated picture and the original image. The findings implied that the generated images might potentially replace the original images if they could be represent the image characteristics of the different glioma subtypes.Conclusion
Standard sequences of superior quality could be generated by using just single T2 sequence. Furthermore, pix2pix outperformed cycleGAN on the same size of dataset for classification of different glioma subtypes.Acknowledgements
This study was supported by the Second Hospital of Lanzhou University-Cuiying Science and Technology Innovation Fund Project (CY2021-BJ-A05).References
1. Conte, G.M.; Weston, A.D.; Vogelsang, D.C.; Philbrick, K.A.; Cai, J.C.; Barbera, M.; Sanvito, F.; Lachance, D.H.; Jenkins, R.B.; Tobin, W.O.; et al. Generative Adversarial Networks to Synthesize Missing T1 and FLAIR MRI Sequences for Use in a Multisequence Brain Tumor Segmentation Model. Radiology 2021, 299, 313-323, doi:10.1148/radiol.2021203786.
2. Sharma A, H.G. Sharma A, Hamarneh G. Missing MRI Pulse Sequence Synthesis Using Multi-Modal Generative Adversarial Network. IEEE Trans Med Imaging 2020, 39, 34-42.
3. Cluceru, J.; Interian, Y.; Phillips, J.J.; Molinaro, A.M.; Luks, T.L.; Alcaide-Leon, P.; Olson, M.P.; Nair, D.; LaFontaine, M.; Shai, A.; et al. Improving the noninvasive classification of glioma genetic subtype with deep learning and diffusion-weighted imaging. Neuro Oncol 2022, 24, 639-652, doi:10.1093/neuonc/noab238.
4. Zhao, J.; Hou, X.; Pan, M.; Zhang, H. Attention-based generative adversarial network in medical imaging: A narrative review. Comput Biol Med 2022, 149, 105948, doi:10.1016/j.compbiomed.2022.105948.
Figures