2119
Prediction of glioma genotypes by APTw-derived radiomic features combined with deep learning networks1Lanzhou University Second Hospital, Lanzhou, China, 2Second Clinical School, Lanzhou University, Lanzhou, China, 3Philips Healthcare, Xi'an, China
Synopsis
Keywords: Tumors, Machine Learning/Artificial Intelligence, Radiomics
The study aimed to predict glioma genotypes combined with radiomic features and deep learning networks by using amide proton transfer (APT) imaging. The genetic subtypes of gliomas can be predicted by radiomics and deep learning networks using conventional MRI, however there are still problems with low accuracy and insufficient generalization. This study puts the screened APT radiomics features into a neural network and compares it with traditional radiomic. The results demonstrated that the proposed model had better performance. Therefore, APTw-derived radiomic features have good ability to predict 3-class molecular typing, providing novel classification tool for non-invasive evaluation for glioma genotypes.Introduction
Clinically, the ability to accurately predict the molecular subtypes of gliomas (IDHmut/1p19qcodel, IDHmut/1p19qnon-codel and IDHwt subtypes) will help to individualize preoperative treatment decisions and predict prognosis. Convolutional neural networks (CNNs) have become widely used in the segmentation, classification, and detection of medical images in recent years[1; 2]. It can deliver a diagnosis that is comparable to or even superior than that of regular doctors by evaluating visual data that cannot be seen by the naked eye. The majority of past studies have used conventional MRI data (e.g. T1WI, T2WI, DWI, etc.) to the extract deep features and put the related metrics into classifiers like SVM, random forest, etc.. However the models that were produced did not generalize effectively[3]. Besides, the common “two-step” classification approach further increases the danger of data overfitting[4]. Amide proton transfer-weighted (APTw) imaging, a non-invasive technique that can evaluate gliomas by detecting changes in the protein concentration at the molecular level[5]. Unlike other studies, we put the APT-derived radiomics features into a CNN network to train a three-class model and compare the outcomes to conventional radiomics.Methods
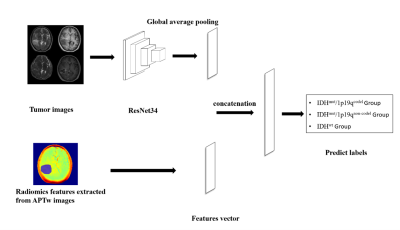
This study included sixty-two patients with diffuse gliomas, dividing into three groups, the IDHmut/1p19qcodel group (22 patients), the IDHmut/1p19qnon-codel group (26 patients) and the IDHwt group (14 patients). All patients underwent MR imaging on a 3T scanner (Ingenia CX, Philips Healthcare, the Netherlands) using a 32 channel Head coil. All images were automatically co-registered to T1WI images by performing a rigid transformation. Regions of interest (ROI) was defined on the areas of abnormal T2-weighted FLAIR signal (including necrosis, cystic degeneration, and edema). PyRadiomics (version 3.9.7) was used to calculate the radiomic features of T2-weighted, T1-weighted sequences before and after administration of a gadolinium-based contrast agent, diffusion-weighted imaging and amide proton transfer-weighted imaging. These included First Order(n=19), Shape-based(n=16), gray-level co-occurrence matrix (GLCM) (n=24), gray-level run length matrix (GLRLM) (n=16), gray-level size zone matrix (GLSZM) (n=16), neighboring gray tone difference matrix (NGTDM) (n = 5), and gray-level dependence matrix (GLDM) (n=14) of original images and a series of transformation derived images based on wavelet transform and Laplacian of Gaussian filter. Within the tumor masks, 1040 radiomics features were collected altogether for each sequence respectively (The total number of features is 1040*4=4160). East absolute shrinkage and selection operator (LASSO) were used for feature selection, and random forest model was used to identify glioma genotypes based on radiomic features (Model A). The sample ratio of training set to test set is 0.7:0.3. Structure of APTw-derived radiomic features combined with deep learning models (Model B) is shown in figure 1. Evaluate model performance by comparing prediction accuracy.Results
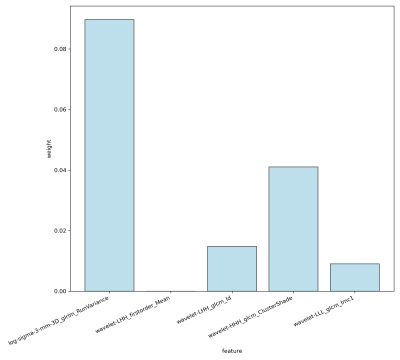
After feature selection, 5 APT features were remained (see figure2). Model B performed well in the test set with an overall accuracy of 90.90% (10/11). Combining APT features with other conventional MRI features, 10 features were get after a new round of screening through LASSO. The Model A classification accuracy in the test sample was 13/19 (68.42%), of which 3/5 (60.00%), 7/10 (70.00%), and 3/4 (75.00%), respectively, were accurate in predicting the IDHmut/1p19qcode, IDHmut/1p19qnon-codel, and IDHwt group. Compared to radiomics alone, combining deep learning networks enhances model performance (Tabel 1).Discussion
In this study we put APT-derived radiological features into Resnet and obtained a better classification performance. A possible explanation for this might be that traditional radiomics mainly focuses on the lesion area and identifies the feature information that cannot be recognized by the human eye, while deep learning does not require manual segmentation and can focus on the entire image. Besides, adding numerical data bridged the transfer learning deficit. Due to the limitation of transfer learning channels, only three different image sequences are contributed to the network for training, which is insufficient to accomplish the goal of multimodal assessment. When the network uses the transfer learning approach, numeric data tries to secure the entry of all sequence information and really implements multi-modal diagnosis. radiomics approaches are used to define numeric data, which is then added to the network structure through fully - connected layers. The combination of the above two provides useful information from different perspectives, which is advantageous for enhancing model performance. This is in line with earlier findings used in glioma grading, which show that the two work best together to increase accuracy[6; 7]. Another possible explanation for the good performance is that APTw images may also provide information on metabolism based on glioma genotype[8]. Furthermore, it is challenging to establish a sample size that is evenly distributed between IDH status and 1p/19q status for a glioma data set. In order to increase accuracy, a two-tiered cascaded approach—first differentiating IDHmut and IDHwt, then differentiating the status of 1p/19q—tends to fit the subtypes with higher data volumes and the three-class model is less likely to overfit and has fewer accumulated errors[9]. Hence, a model for APT-derived radiological signatures combined with Resnet34 can provide a promising way to differentiate gliomas genotype and help to give an additional imaging evidence to clinical diagnosis.Conclusion
APT-derived radiomic signature combined with 3-class Resnet34 has higher classification accuracy, which may provide a reliable classification tool for non-invasive assessment of glioma genotype.Acknowledgements
This study was supported by the Second Hospital of Lanzhou University-Cuiying Science and Technology Innovation Fund Project (CY2021-BJ-A05).References
1 Chang K, Beers AL, Bai HX et al (2019) Automatic assessment of glioma burden: a deep learning algorithm for fully automated volumetric and bidimensional measurement. Neuro Oncol 21:1412-1422
2 Jin L, Shi F, Chun Q et al (2021) Artificial intelligence neuropathologist for glioma classification using deep learning on hematoxylin and eosin stained slide images and molecular markers. Neuro Oncol 23:44-52
3 Ding J, Zhao R, Qiu Q et al (2022) Developing and validating a deep learning and radiomic model for glioma grading using multiplanar reconstructed magnetic resonance contrast-enhanced T1-weighted imaging: a robust, multi-institutional study. Quant Imaging Med Surg 12:1517-1528
4 Cluceru J, Interian Y, Phillips JJ et al (2022) Improving the noninvasive classification of glioma genetic subtype with deep learning and diffusion-weighted imaging. Neuro Oncol 24:639-652
5 Zhou J, Lal B, Wilson DA, Laterra J, van Zijl PC (2003) Amide proton transfer (APT) contrast for imaging of brain tumors. Magn Reson Med 50:1120-1126
6 Yang Y, Yan LF, Zhang X et al (2018) Glioma Grading on Conventional MR Images: A Deep Learning Study With Transfer Learning. Front Neurosci 12:804
7 Zhang Z, Xiao J, Wu S et al (2020) Deep Convolutional Radiomic Features on Diffusion Tensor Images for Classification of Glioma Grades. J Digit Imaging 33:826-837
8 Reitman ZJ, Jin G, Karoly ED et al (2011) Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proceedings of the National Academy of Sciences 108:3270-3275
9 Matsui Y, Maruyama T, Nitta M et al (2020) Prediction of lower-grade glioma molecular subtypes using deep learning. J Neurooncol 146:321-327
Figures