2116
Radiomics and Machine Learning for Prediction of Relapsed and Refractory Primary Central Nervous System Lymphoma1Department of Radiological Sciences, University of California, Irvine, CA, United States, 2Department of Medical Imaging, Chi Mei Medical Center, Tainan, Taiwan, 3Department of Health and Nutrition, Chia Nan University of Pharmacy and Science, Tainan, Taiwan, 4Department of Radiology, E-DA Hospital, I-Shou University, Kaohsiung, Taiwan
Synopsis
Keywords: Tumors, Machine Learning/Artificial Intelligence
A subset of primary central nervous system lymphoma (PCNSL) may show early relapsed/refractory (R/R) disease after treatments. This study investigated the role of radiomics and machine learning for the prediction of R/R in PCNSL after treatments. Total 46 patients with pathologically confirmed PCNSL were included. Total 321 radiomic features were extracted from various pre-treatment MR sequences in each patient to build prediction models. Among various machine learning algorithms, the best predictive performance with accuracy of 82.6%, precision of 80%, and AUC of 0.85 were obtained in support vector machine.Background and Purpose
Primary central nervous system lymphoma (PCNSL), a rare subtype of non-Hodgkin’s lymphoma (NHL), accounts for 3-4% of central nervous system (CNS) tumors and 1-2% of NHLs [1,2]. Diffuse large B- cell lymphoma (DLBCL) is the most common subtype of PCNSL [3]. Currently, there is no optimal standard salvage regimen. High-dose (HD) methotrexate (MTX)-based chemotherapy is the cornerstone of therapy in PCNSL, while performing whole-brain radiotherapy (WBRT) after chemotherapy yields a better survival rate than chemotherapy alone [4,5]. Although MTX-based chemotherapy and WBRT have been considered to be effective therapies for PCNSL [4,5], up to 50-60% of patients eventually had relapsed/refractory (R/R) disease, and the prognosis in R/R PCNSL remains poor [6,7]. Conventional MRI findings such as tumor size, infratentorial localization, and non-enhancing T2-fluid attenuated inversion recovery (FLAIR) hypersignal lesion have been reported as important variables related to prognosis in PCNSL [8]. However, quantitative radiomics analysis for prediction of R/R in PCNSL is rare. Thus, we investigated the role of radiomics and machine learning for the prediction of R/R in PCNSL in this study.Materials and Methods
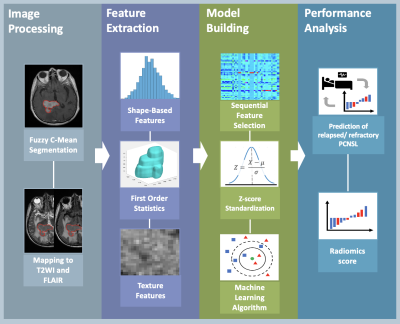
The MRI dataset acquired from two venders (Siemens and GE Healthcare) were used to build radiomics model. Total 46 PCNSL patients with complete pre-treatment brain MRI and post-treatment MRI follow up were included, including 20 patients with R/R and 26 patients with non-R/R disease. All patients received HD-MTX based chemotherapy. For each patient, the lesion region of interest (ROI) was delineated on contrast-enhanced T1-weighted imaging (CE-T1WI) by using Fuzzy C-Means (FCM) Clustering algorithm [9]. To ensure the ROI to be corresponded to all MRI sequences, the segmented ROI was co-registered to T2-weighted imaging (T2WI) and fluid-attenuation inversion recovery (FLAIR) by using the open-source ITK-SNAP 4.0. Radiomic features were extracted from CE-T1WI, T2WI, and FLAIR using pyradiomics 3.0 in python [10]. A total of 107 radiomics features were calculated for each MR sequence, including 14 shape-based features, 18 first-order statistics features, and 75 texture features. To evaluate the importance of extracted features for predicting R/R in PCNSL, the support vector machine (SVM) with gaussian kernel method were used to select feature [11]. Various machine learning algorithms were used to build radiomic models with 10-fold cross-validation, including SVM, k-nearest neighbors (KNN), and ensemble learning algorithms. Moreover, the radiomics score was calculated for each patient to predict R/R probability in PCNSL. Figure 1 shows the flowchart in establishing the radiomics-based predictive models. All procedure was implemented in MATLAB 2022 [12].Results
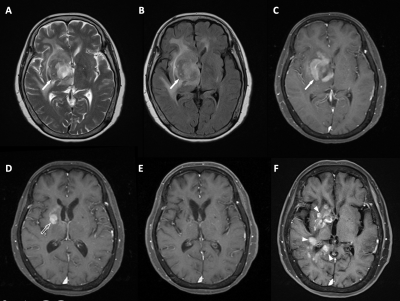
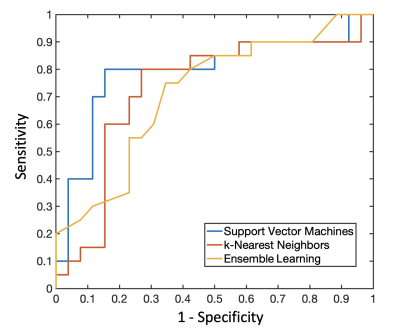
Of the included 46 PCNSL patients, 20 (20/46, 43.5%) patients were found to have R/R (Figure 2), and 26 (26/46, 56.5%) patients remained non-R/R. The most significant five radiomic features were selected by the SVM classifier for differentiation of R/R. Among prediction models, SVM was superior to KNN and ensemble learning for prediction of R/R in PCNSL. The predictive results in SVM showed 16 true positive cases, 22 true negative cases, 4 false positive cases, and 4 false negative cases. The overall prediction accuracy of 82.6%, precision of 80%, and AUC of 0.85 in SVM was obtained. The receiver operating characteristic (ROC) curves for all three models were shown in Figure 3.Conclusions
This study attempted to use radiomics and machine learning for the prediction of R/R in PCNSL after treatments. Our results showed that radiomic analysis based on pre-treatment brain MRI, including CE-T1WI, T2WI, and FLAIR may provide valuable information for treatment planning in PCNSL.Acknowledgements
No acknowledgement found.References
1. Hoffman, S.; Propp, J.M.; McCarthy, B.J. Temporal trends in incidence of primary brain tumors in the United States, 1985-1999. Neuro-oncology 2006, 8, 27-37, doi:10.1215/s1522851705000323.
2. Villano, J.L.; Koshy, M.; Shaikh, H.; Dolecek, T.A.; McCarthy, B.J. Age, gender, and racial differences in incidence and survival in primary CNS lymphoma. British journal of cancer 2011, 105, 1414-1418, doi:10.1038/bjc.2011.357.
3. Montesinos-Rongen, M.; Brunn, A.; Bentink, S.; Basso, K.; Lim, W.K.; Klapper, W.; Schaller, C.; Reifenberger, G.; Rubenstein, J.; Wiestler, O.D.; et al. Gene expression profiling suggests primary central nervous system lymphomas to be derived from a late germinal center B cell. Leukemia 2008, 22, 400-405, doi:10.1038/sj.leu.2405019.
4. Morris, P.G.; Abrey, L.E. Therapeutic challenges in primary CNS lymphoma. Lancet Neurol 2009, 8, 581-592, doi:10.1016/s1474-4422(09)70091-2.
5. Song, J.; Samant, R.; Jay, M.; Chaudry, H.; Fan, X.Y.; MacDonald, D.; Bence-Bruckler, I.; Nair, V. Whole brain radiotherapy improves survival outcomes in primary CNS lymphoma patients ineligible for systemic therapy. Supportive Care in Cancer 2020, 28, 5363-5369, doi:10.1007/s00520-020-05376-2.
6. Grommes, C.; DeAngelis, L.M. Primary CNS Lymphoma. Journal of Clinical Oncology 2017, 35, 2410-2418, doi:10.1200/jco.2017.72.7602.
7. Korfel, A.; Schlegel, U.; Herrlinger, U.; Dreyling, M.; Schmidt, C.; von Baumgarten, L.; Pezzutto, A.; Grobosch, T.; Kebir, S.; Thiel, E.; et al. Phase II Trial of Temsirolimus for Relapsed/Refractory Primary CNS Lymphoma. J Clin Oncol 2016, 34, 1757-1763, doi:10.1200/jco.2015.64.9897.
8. Tabouret, E.; Houillier, C.; Martin-Duverneuil, N.; Blonski, M.; Soussain, C.; Ghesquières, H.; Houot, R.; Larrieu, D.; Soubeyran, P.; Gressin, R.; et al. Patterns of response and relapse in primary CNS lymphomas after first-line chemotherapy: imaging analysis of the ANOCEF-GOELAMS prospective randomized trial. Neuro-oncology 2017, 19, 422-429, doi:10.1093/neuonc/now238.
9. Nie, K.; Chen, J.H.; Yu, H.J.; Chu, Y.; Nalcioglu, O.; Su, M.Y. Quantitative analysis of lesion morphology and texture features for diagnostic prediction in breast MRI. Acad Radiol 2008, 15, 1513-1525, doi:10.1016/j.acra.2008.06.005.
10. van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Research 2017, 77, e104-e107, doi:10.1158/0008-5472.Can-17-0339.
11. Tong, S.; Chang, E. Support vector machine active learning for image retrieval. In Proceedings of the Proceedings of the ninth ACM international conference on Multimedia, Ottawa, Canada, 2001; pp. 107–118.
12. MATLAB. Version 9.13.0 (R2022b). Natick, Massachusetts: The MathWorks Inc.; 2022.
Figures