2068
mpMRI Radiomic Features Predict the Likelihood for Progression to Treatment of Prostate Cancer Patients on Active Surveillance1Department of Radiation Oncology, University of Miami Miller School of Medicine, Miami, FL, United States, 2Department of Urology, University of Miami Miller School of Medicine, Miami, FL, United States, 3Department of Radiology, University of Miami Miller School of Medicine, Miami, FL, United States, 4Department of Pathology, University of Miami Miller School of Medicine, Miami, FL, United States
Synopsis
Keywords: Quantitative Imaging, Prostate, Active Surveillance, mpMRI, Prostate Cancer
Active surveillance (AS) for prostate cancer has emerged as a safe and attractive alternative to immediate treatment. Here we present an integrated method for baseline mpMRI analysis enabling early detection of patients harboring lesions with a high potential for progression. The approach consists of three steps: (i) Training a deep learning network for automatic segmentation of prostate and lesions, suspicious for cancer; (ii) Application of the network to identify lesions on mpMRI images for patients, enrolled in an AS trial; and (iii) Development of a progression risk stratification model by incorporating radiomic and clinical variables.Purpose
Active surveillance (AS) has emerged as a safe and attractive alternative to immediate treatment and has been incorporated into many prostate cancer management guidelines.1-3 However, missing the window for cure is still a concern, as emerging data show an increased risk of metastasis with long follow-up.4 There is an unmet clinical need in AS for developing robust baseline risk-stratification tools enabling early detection of patients harboring lesions with a high potential for progression. Early identification of patients who are not good candidates for AS will help avoiding delays in the initiation of definitive therapy. Prostate cancer multifocality and heterogeneity5 are the Achilles' heel of prostate cancer risk stratification. Multiparametric MRI (mpMRI) provides a promising glimpse into tumor heterogeneity6 and here we present an approach for predicting the likelihood of progression based on baseline mpMRI and clinical variables for patients, enrolled in AS. The resulting model aims to improve the prospective AS patient selection.Materials and methods
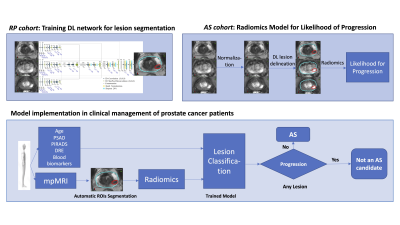
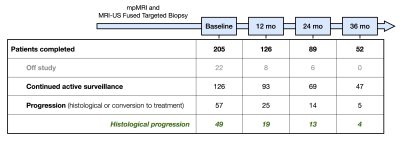
The approach consists of three steps depicted in Figure 1: (i) Training a deep learning (DL) network for automatic segmentation of prostate and lesions, suspicious for cancer; (ii) Application of the network to identify lesions on mpMRI for patients, enrolled in AS; and (iii) Radiomic feature extraction and model building for prediction of patients who will progress. First, a 3D nnU-Net7 was trained on mpMRI from patients undergone radical prostatectomy (RP). This was performed as a two-stage process, where the prostate was first segmented and introduced to the lesion detection network. The prostate contours were manually outlined in MIM (MIM software, Cleveland, Ohio). The pathologist’s outlined lesions were translated onto T2-weighted (T2W) sequence as previously described.8 The high B-value (BVAL), ADC, the early enhancing series from DCE-MRI and T2W images were used in the training. Second, the network was applied to identify lesions on baseline mpMRI of patients, participating in a single arm AS trial “MRI-Guided Biopsy Selection of Prostate Cancer Patients for Active Surveillance versus Treatment: The Miami MAST Trial” (ClinicalTrials.gov: NCT02242773). Upon consent, patients undergo mpMRIs exams at baseline and during 12-month, 24-month and 36-month follow-up visits. Using the trained nnU-Net, the lesions were automatically detected on the baseline mpMRI. A series of radiomic features were extracted from the ADC, T2W, BVAL and DCE sequences for each lesion that included lesion volume, intensity mean, standard deviation, skewness, kurtosis and 10/25/50/75/90 percentiles. On a patient level, the minimum, maximum and mean of lesion-based features across all detected lesions were computed for each of the four MRI sequences. In addition, the total detected prostate volume, total combined lesion volume and total lesion count were added to the model, amounting to a total of 114 summary features per patient. A minimum Redundancy - Maximum Relevance (mRMR)8 feature selection method was used to rank the features with histological progression of the MAST trial patients as the categorical target variable. Histological progression was defined as: (i) more than 4 positive cores involving any grade of cancer, (ii) three or more cores with Gleason 3+4 cancer, (iii) any single core with Gleason 4+3 cancer or higher. An additional logistic regression based cross validated exhaustive feature search was applied to select the most significant top ranked features to maximize the area under the receiver operating characteristic curve (ROC-AUC). Clinical features, i.e. age, body mass index (BMI), PSA density, maximum PI-RADS score, blood biomarker 4Kscore and NCCN risk classification, were also added to the model. A logistic regression model was built using the selected features to predict patients with high risk of progression to treatment.Results
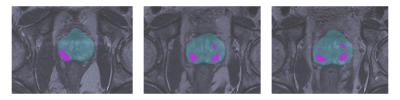
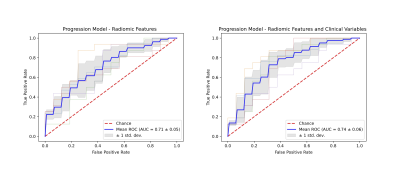
The nnU-Net was trained with 45 mpMRIs from RP patients with a total of 174 lesions using 5-fold cross validation, resulting in a mean accuracy of 99.7% for prostate segmentation and 94.7% for lesion detection. Running inference on the 205 patients of the MAST trial resulted in at least one lesion being detected in 96.6% of the patients (Figure 2). Seven features were selected for the final model. A total of 161 patients were analyzed, out of which 81 showed histological progression at some point during the MAST trial (Figure 3, excluding patients for which the nnU-Net did not identify any lesions). The final radiomics model achieved a 5-fold cross validation mean AUC of 0.71. After adding clinical variables to the radiomics model the mean AUC increased to 0.74 (Figure 4).Discussion
Image-based automated prostate cancer classification is an area of active investigation. The standard underlying schema of the approach consists of feature extraction and classification. Here we investigated the ability of a radiomics model to predict the likelihood of progression for patients enrolled in AS management. We designed an automatic procedure, whereby the prostate and prostate cancer lesions are automatically delineated and a comprehensive radiomics pipeline is used to extract a series of quantitative features from all sequences of mpMRI. The resultant model has sensible performance. Further training of the nnU-Net with more data will most likely result in improved performance.Conclusions
Our analysis indicates that there is a predictive signal in the radiomics variables related to progression, and our model can prospectively improve AS patient selection. To our knowledge, this is the first work that tackles this important problem.Acknowledgements
No acknowledgement found.References
1. Wilt TJ, Brawer MK, Jones KM, et al. Radical prostatectomy versus observation for localized prostate cancer. N Engl J Med. 2012;367:203-213. DOI: 10.1056/NEJMoa1113162.
2. Hamdy FC, Donovan JL, Lane JA, et al. 10-Year Outcomes after Monitoring, Surgery, or Radiotherapy for Localized Prostate Cancer. N Engl J Med. 2016;375:1415-1424. DOI: 10.1056/NEJMoa1606220.
3. Klotz L, Zhang L, Lam A, et al. Clinical results of long-term follow-up of a large, active surveillance cohort with localized prostate cancer. J Clin Oncol 28. 2010;28(1):126-131. DOI: 10.1200/JCO.2009.24.2180.
4. Yamamoto T, Bindu Musunuru H, Vesprini D, et al. Metastatic Prostate Cancer in Men Initially Treated with Active Surveillance. J Urol. 2016;195(5):1409-1414. DOI: 10.1016/j.juro.2015.11.075.
5. Punnen S, Stoyanova R, Kwon D, et al. Heterogeneity in Genomic Risk Assessment From Tissue Based Prognostic Signatures Used in the Biopsy Setting and the Impact of MRI Targeted Biopsy. J Urol. 2021;205(5):1344-1351. DOI: 10.1097/JU.0000000000001559.
6. Chang YCC, Ackerstaff E, Tschudi Y, et al. Delineation of Tumor Habitats based on Dynamic Contrast Enhanced MRI. Sci Rep. 2017;7:9746. DOI: 10.1038/s41598-017-09932-5.
7. Isensee F, Jaeger PF, Kohl SAA, et al. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods. 2021;18:203–211. DOI: 10.1038/s41592-020-01008-z.
8. Peng H, Long F, Ding C. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intell. 2005; 27(8):1226-1238. DOI: 10.1109/TPAMI.2005.159.
Figures