2058
Kaplan-Meier survival analysis of the DCE-MRI-based radiomics in the prognosis of prostate cancer1Clinical medicine school of Ningxia Medical University, Yinchuan, China, 2Department of Radiology, the First Hospital Affiliated to Hainan Medical College, Haikou, China, 3GE Healthcare, Beijing, China, 4Department of Radiology, General Hospital of Ningxia Medical University, Yinchuan, China
Synopsis
Keywords: Prostate, DSC & DCE Perfusion
In this study, we aim to investigate whether DCE-MRI-based radiomics can be used in the prognosis of prostate cancer. Rad-score in which DEC-MRI-based radiomics labels were established by screening radiomics features GLCM_Correlation and shape_Flatness through Lasso regression was significantly different between the death group and the censor group. It was concluded that DCE-MRI-based radiomics can be used as prognostic factors for prostate cancer.Summary of Main Findings
DCE-MRI-based radiomics can be used as prognostic factors for prostate cancer. Rad-score was significantly different among the death group and the censor group.Introduction
Prostate cancer (PCa) is the second most common malignant cancers, and fifth leading cause of cancer death among men in 20201-2. In recent years, the incidence rate of PCa continue to increase in China, ranking sixth in incidence and seventh in mortality3. Currently, in clinical practice, radiologists evaluate traditional qualitative features of PCa such as density, enhancement, margin, surrounding tissues and structures4. However, medical imaging has much richer information that the human eye can’t distinguish5. Radiomics can extract and analyze quantitative features from medical imaging which information obtained can be applied to support the clinical decision, diagnosis, treatment, and prognosis6, and have successfully applied in lung cancer, thyroid cancer, breast cancer et al7-9. Magnetic resonance imaging (MRI) is widely used for PCa diagnosis, treatment and has proven a promising and efficient modality for PCa10. The development of radiomics has provided a new dimension of MRI analysis, a more impartial and disinterested way via MRI quantity feature extraction. Previous studies have reported positive results on MRI-based radiomics applied in PCa detection, classification of grade groups, and Gleason score prediction et al11. This study aims to investigate feasibility of applying DCE-MRI-based radiomics features in the prognosis of PCa through Kaplan-Meier survival analysis.Material and Methods
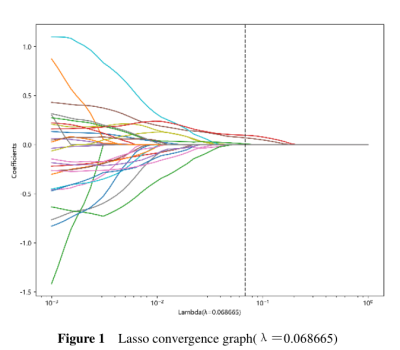
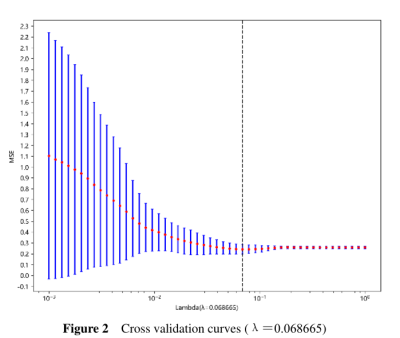
We retrospectively collected 76 patients who were pathologically diagnosed PCa in the general hospital of Ningxia Medical University from Jan 1, 2008, to Dec 31, 2014. All cases were followed up. The starting point of the follow-up was the patient's admission date, and the deadline was December 31, 2021. The data of each patient was visually graded by a radiologist and a pathologist. All patients underwent an MRI examination before biopsy and operation (GE Signa Excite HD 3.0 T, USA). In addition, some parts of the patient’s clinical, and laboratory data were recorded from the health information system (HIS). 3D slicer 5.1.0 (https://www.slicer.org) software was used to delineate volume of interest on DCE-MRI based PCa images. For radiomics feature extraction, Python Anaconda (3.0) was used to extract high-throughput image features of the PCa. For features with high repeatability (ICC > 0.9), one of the two features should be retained. We used greedy recursive deletion strategy for feature filtering. The LASSO regression model was used on the discovery data set for signature construction. To find an optimal ‘λ’, 10-fold cross validation with minimum criteria was employed, where the final value of ‘λ’ yielded minimum cross validation error. The retained features with nonzero coefficients were used for regression model fitting and combined into a radiomics signature. Subsequently, we obtained a radiomics score for each patient by a linear combination of retained features weighed by their model coefficients. The Python scikit-learn package was used for LASSO regression modeling. All cases were followed-up. A two‑tailed P‑value of < 0.05 was considered to indicate a statistically significant difference.Results
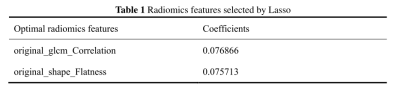
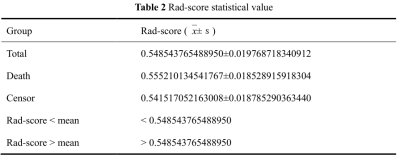
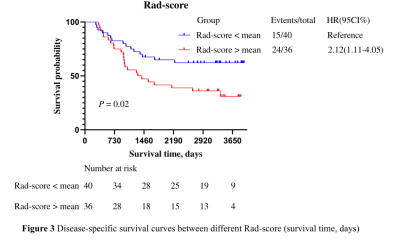
PCa patients’ 1, 3, 5 years disease-specific survival rates were 93. 4%, 69.7%, and 53.9% respectively. The median survival time was 2557 days. We extracted 107 radiomics features, including 14 (13.1%) shape features, 24 (22.4%) Gray-Level Co-occurrence Matrix (GLCM), 14 (13.1%) Gray Level Dependence Matrix (GLDM), 16 (15.0%) Gray Level Run Length Matrix (GLRLM), 16 (15.0%) Gray Level Size Zone Matrix (GLSZM), 5 (4.7%) Neighborhood Grey Tone Difference Matrix (NGTDM ), 18 (16.8%) First-order features. After Spearman's rank correlation, 28 features were kept. Final value of λ=0.068665 yielded minimum cross validation error, the retained nonzero 2 features were kept to construct Rad-signature (Figure 1, Figure 2). Subsequently, we obtained a radiomics score for each patient by a linear combination of retained features weighed by their model coefficients. Rad-score = 0.479377342821061 + 0.076866 * original_glcm_Correlation + 0.075713 * original_shape_Flatness. Mean Rad-score = 0.548543765488950±0.019768718340912(Table 1). According to the mean Rad-score, the patients were divided into < mean Rad-score group and > mean Rad-score group (Table 2). Kaplan-Meier was performed to analyze DCE-MRI-based radiomics in the prognosis of PCa. The results showed that there were statistically significant between the two groups (χ2 = 5.42, 95%CI:1.11-4.05 P = 0.02). The > mean Rad-score group mortality risk was 2.12 times that of the < mean Rad-score group (Figure 3).Discussion and Conclusion
Our study demonstrated that Rad-score which DEC-MRI-based radiomics labels were established by screening radiomics features GLCM_Correlation and shape_Flatness through Lasso regression was associated with the prognosis of PCa. Previous articles have reported that GLCM_Correlation is correlated with metastasis-free survival of PCa12. GLCM_Correlatio can also predict hepatic metastasis of pancreatic ductal carcinoma13. Other articles reported that shape_Flatness can predict Ki-67 expression in adrenocortical carcinoma14. Shape_flatness can also be used to predict the good pathological response of non-small cell lung cancer patients receiving immunotherapy-based neoadjuvant therapy15. Our results were in agreement with those previous studies in terms of GLCM_Correlation and shape_Flatness established radiomics labels correlate with patient outcomes. Furthermore, DCE-MRI-based radiomics can predict the prognosis of PCa, hence help with better risk stratification and precision medicine for patients. However, future prospective study with a bigger cohort of investigation is warranted. To conclude, DCE-MRI-based radiomics can be used as prognostic factors for PCa.Acknowledgements
This study was supported by grants from The Key Research and Development Program of Ningxia (No.2019BEG03033) and Natural Science Foundation of Ningxia(2022AAC03472). Author contributions: conception and design: Ting Huang, Zhiqiang Chen, Shaoru Zhang, Zhuo Wang, Xiaohua Chen, Yunshu Zhou,Xiaocheng Wei, AiJun Wang; Data analysis and interpretation: TingHuang, Zhiqiang Chen, Shaoru Zhang, Zhuo Wang, Xiaohua Chen, Yunshu Zhou,Xiaocheng Wei, AiJun Wang; drafting the article and revising it critically for important intellectual content: Zhiqiang Chen; final approval of manuscript: all authors. The authors declare no conflicts of interest.References
1. Sung H, Ferlay J, Siegel R L, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries[J]. CA Cancer J Clin, 2021, 71(3): 209-249.
2. Culp M B, Soerjomataram I, Efstathiou J A, et al. Recent global patterns in prostate cancer incidence and mortality rates[J]. Eur Urol, 2020, 77(1): 38-52.
3. Cao W, Chen H D, Yu Y W, et al. Changing profiles of cancer burden worldwide and in China: A secondary analysis of the global cancer statistics 2020[J]. Chin Med J (Engl), 2021, 134(7): 783-791.
4. Bi W L, Hosny A, Schabath M B, et al. Artificial intelligence in cancer imaging: Clinical challenges and applications[J]. CA Cancer J Clin, 2019, 69(2): 127-157.
5. Avanzo M, Wei L, Stancanello J, et al. Machine and deep learning methods for radiomics[J]. Med Phys, 2020, 47(5): 185-202.
6. Scapicchio C, Gabelloni M, Barucci A, et al. A deep look into radiomics[J]. Radiol Med, 2021, 126(10): 1296-1311.
7. Shimada Y, Kudo Y, Maehara S, et al. ASO visual abstract: Radiomics with artificial intelligence for the prediction of early recurrence in patients with clinical stage IA lung cancer[J]. Ann Surg Oncol, 2022.
8. Sorrenti S, Dolcetti V, Radzina M, et al. Artificial intelligence for thyroid nodule characterization: Where are we standing? [J]. Cancers (Basel), 2022, 14(14).
9. Fang C, Zhang J, Li J, et al. Clinical-radiomics nomogram for identifying HER2 status in patients with breast cancer: A multicenter study[J]. Front Oncol, 2022, 12: 922185.
10. El K C, Ros P R. A systematic review for health disparities and inequities in multiparametric magnetic resonance imaging for prostate cancer diagnosis[J]. Acad Radiol, 2021, 28(7): 953-962.
11. Spohn S, Bettermann A S, Bamberg F, et al. Radiomics in prostate cancer imaging for a personalized treatment approach - current aspe cts of methodology and a systematic review on validated studies[J]. Theranostics, 2021, 11(16): 8027-8042.
12. Franzese C, Cozzi L, Badalamenti M, et al. Radiomics-based prognosis classification for high-risk prostate cancer treated with radiotherapy[J]. Strahlenther Onkol, 2022, 198(8): 710-718.
13. De Robertis R, Geraci L, Tomaiuolo L, et al. Liver metastases in pancreatic ductal adenocarcinoma: A predictive model based on CT texture analysis[J]. Radiol Med, 2022, 127(10): 1079-1084.
14. Ahmed A A, Elmohr M M, Fuentes D, et al. Radiomic mapping model for prediction of Ki-67 expression in adrenocortical carcinoma[J]. Clin Radiol, 2020, 75(6): 417-479.
15. Lin Q, Wu H J, Song Q S, et al. CT-based radiomics in predicting pathological response in non-small cell lung cancer patients receiving neoadjuvant immunotherapy[J]. Front Oncol, 2022, 12: 937277.