2036
Automatic Couinaud segmentation in Intravenous-Phase Enhanced MRI images by key point detection with deep neural network1Chengdu Institute of Computer Application, Chinese Academy of Sciences, Chengdu, China, 2School of Computer Science and Technology, University of Chinese Academy of Sciences, Beijing, China, 3Department of Radiology, First Affiliated Hospital of Dalian Medical University, Dalian, China, 4Dalian Engineering Research Center for Artificial Intelligence in Medical Imaging, Dalian, China
Synopsis
Keywords: Liver, Liver, Couinaud segmentation
This study presents an effective framework for automatic Couinaud liver segmentation in Intravenous-Phase Enhanced MRI images, without the time-consuming delineation of each segment or identifying different branches of the vessel system. We define seven key points located at the bifurcations of the vascular system to divide liver into Couinaud segments II-VIII. We train a key point detection model to locate the coordinate of key point. The overall dice score is 81.17% and average surface distance is 2.35mm in test set.
Introduction
The couinaud segmentation system1, which divides the liver into eight functionally independent segments based on the bifurcation of hepatic veins and portal veins, plays a significant role in liver surgical planning and lesion monitoring2. However, due to the intricacy of the intrahepatic vascular system, automated Couinaud segmentation has been a challenging problem. To tackle this issue, we build a deep learning-based key point detection model for Couinaud segments II-VIII extraction.Methods
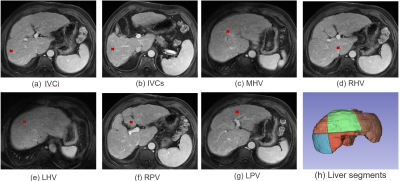
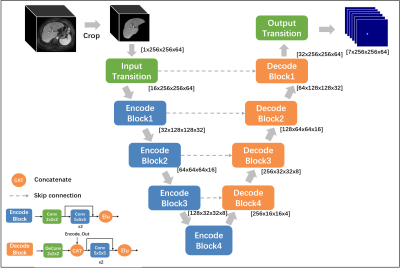
As shown in Figure 1, we define 7 key points located at the bifurcations of the vascular system including 1) Inferior vena cava, inferior part(IVCi); 2) Inferior vena cava, superior part(IVCs); 3) Middle hepatic vein(MHV); 4) Right hepatic vein(RHV); 5) Left hepatic vein(LHV); 6) Right portal vein(RPV); 7) Left portal vein(LPV), the liver is divided into segments II-VIII using the plane built by 7 points using predefined rules3.To detect the coordinates of above mentioned seven key points, we transfer each point into a heat map and built a 3D MRI segmentation model. As shown in Figure 2, we use a typical semantic segmentation model VNet4 as our backbone. The input is first cropped according to its liver mask to discard irrelevant information, then we use VNet to infer the location of seven key points. The network output is a seven-channel heat map at the same resolution as the cropped liver image. The Input transition and Output transition layers are composed of a 5x5x5 convolution layer to map the channel to the hidden dim(16) and the number of key points (7). Each Encode block halves the feature resolution as well as doubles the number of channels; On the opposite, each decoding block doubles the resolution, combines it with the encoder output of the same level and halves the number of channels.
We use Kullback-Leibler(KL) divergence loss as our loss function.
Results
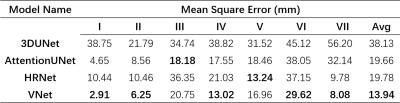
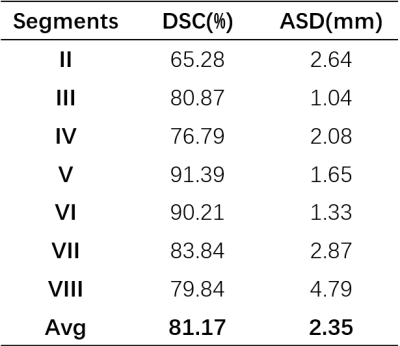
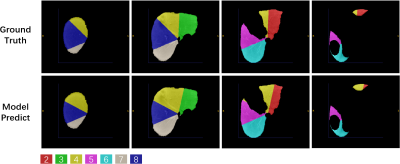
Dataset: We collected 50 Hepatocellular carcinoma patients in The First Affiliated Hospital of Dalian Medical University, each case has an Intravenous-Phase Enhanced MRI image and corresponding delineation labels of the liver and seven vascular bifurcation points. All labels are manually delineated by an experienced physician and confirmed by two senior physicians. In the experiment setting, 50 MRI images were divided into train/validation/test/ splits with the ratio of 8:1:1 and used 5-fold cross-validation to evaluate our model.We quantitatively compare three semantic segmentation models including 3DUNet5, AttentionUNet6 and VNet4 and one landmark detection model named HRNet7 with the Mean Square Error(MSE) in the test set. The results are shown in Table.1. VNet performs best with an average MSE of 13.94 mm. Following, we extract segments II-VIII with the predicted keypoint coordinate and compute the dice score(DSC) and average surface distance(ASD) with the Ground-truth generated by the labelled key point. The results are shown in Table.2. Our model performs acceptable results with DSC 81.17% and ASD 2.35 mm. Figure.3 shows the ground truth and predicted segments. The predicted segmentation shows our framework can effectively extract Couinaud segments in MRI images, the black hollow indicates tumour areas.
Discussion
We trained a deep learning-based keypoint detection network to locate seven key points distributed at the bifurcations of the vascular system and divide the liver into segments II-VIII automatically. The mean square error of our detection network is 13.94 mm, and the dice score and average surface distance of divided segments are 81.17% and 2.35 mm. Nevertheless, one limitation of our approach is that segment I cannot be derived from key points. For complete automatic Couinaud segmentation, we need a separate semantic segmentation model to get segment I.Conclusion
This study has presented an effective framework for automatic Couinaud liver segmentation in Intravenous-Phase Enhanced MRI images, without the time-consuming delineation of each segment or identifying different branches of the vessel system. Our framework can effectively divide the liver into independent units and intuitively display the corresponding relationship between liver segments and the tumour areas, thus better-assisting liver surgery planning and lesion monitoring.Acknowledgements
First I would like to thank my partner Ren Xue from Dalian Medical University for this work, we discussed closely with each other and overcame the problems we encountered; I would also like to thank Prof Liu Ailian and Dr Zhao Ying from Dalian Medical University for their careful guidance. Finally, I would like to express my gratitude to my supervisor, Prof Yao Yu, and other members of the research group.References
[1] C. Couinaud, Liver anatomy: portal (and Suprahepatic) or biliary segmentation, Digest. Surg. 16 (6) (1999) 459–467, https://doi.org/10.1159/000018770.
[2] H. Bismuth, Surgical anatomy and anatomical surgery of the liver, World J. Surg. 6 (1) (Jan. 1982) 3–9, https://doi.org/10.1007/BF01656368.
[3] Germain, T. et al. Liver segmentation: Practical tips. Diagnostic and Interventional Imaging 95, 1003–1016 (2014).
[4] Milletari, F., Navab, N. & Ahmadi, S.-A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. Preprint at https://doi.org/10.48550/arXiv.1606.04797 (2016).
[5]Çiçek, Ö., Abdulkadir, A., Lienkamp, S. S., Brox, T. & Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. Preprint at https://doi.org/10.48550/arXiv.1606.06650 (2016).
[6]Oktay, O. et al. Attention U-Net: Learning Where to Look for the Pancreas. Preprint at https://doi.org/10.48550/arXiv.1804.03999 (2018).
[7]Sun, K. et al. High-Resolution Representations for Labeling Pixels and Regions. Preprint at https://doi.org/10.48550/arXiv.1904.04514 (2019).
Figures