2019
Pulseq-gSlider: Scanner-independent high-isotropic-resolution dMRI on an open-source platform1Department of Psychiatry, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA, United States, 2School of Biomedical Engineering, Southern Medical University, Guangzhou, China, 3Department of Radiology, Stanford University, Stanford, CA, United States, 4Department of Electrical Engineering, Stanford University, Stanford, CA, United States, 5Institute of Imaging Science, Department of Biomedical Engineering, Department of Electrical Engineering, Vanderbilt University, Nashville, TN, United States, 6Functional MRI Laboratory, University of Michigan, Ann Arbor, MI, United States, 7Fetal-Neonatal Neuroimaging and Developmental Science Center, Boston Children’s Hospital, Harvard Medical School, Boston, MA, United States, 8Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA, United States, 9Department of Radiology, Harvard Medical School, Cambridge, MA, United States, 10Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA, United States, 11Department of Radiology, Medical Physics, University Medical Center Freiburg, Freiburg, Germany, 12Department of Biomedical Engineering, University of Michigan, Ann Arbor, MI, United States, 13Department of Radiology, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA, United States
Synopsis
Keywords: Data Acquisition, Software Tools
Using on the open-source pulse sequence programming platform Pulseq, we successfully developed and implemented the gSlider sequence on a 3T scanner without the vendor-specific sequence development environment. One-millimeter whole-brain isotropic dMRI data was acquired using the gSlider implementation from Pulseq and Siemens and test-retest variability was evaluated. The test-retest reliability in fractional anisotropy improved dramatically for the Pulseq implementation while it was comparable for both implementations for mean diffusivity. We demonstrated the feasibility of implementing advanced sequences using the open-source Pulseq platform and expect that it will be easily transferred to and executed on different vendors.Introduction
Diffusion MRI (dMRI) plays a key role in understanding the microstructure and function of the human brain. In recent years, advanced sequences which pursue high-isotropic resolution dMRI have been studied and developed1–3. One representative work is Generalized SLIce Dithered Enhanced Resolution (gSlider)4,5, which uses RF pulses with different subslice profiles and provides high SNR-efficiency. However, implementing such sequences in vendor-specific environments is extremely resource and time intensive and impedes their ability to share across sites and vendors, thereby hindering scientific progress.Motivated by making designing and implementing cutting-edge pulse sequences flexible, and more importantly, to harmonize quantitative MRI studies across multiple collaborative sites, open-source pulse programming platforms Pulseq6 and TOPPE7 provide a new file format to describe sequence components in a human-readable, shareable, and compact way. Within one file, sequence instructions such as RF pulses, gradients, ADC events, and delays are programmed. Using vendor-specific interpreters, the generated seq file can be executed on the scanners.
In the present work, with the support of the vendor-independent sequence design ability of the Pulseq platform, we developed and implemented the gSlider sequence. One-millimeter isotropic resolution whole-brain dMRI was acquired on a 3T Siemens Prisma scanner. We compared the performance of our Pulseq-gSlider sequence with the vendor-specific gSlider sequence. We also assessed the test-retest reliability of both gSlider sequences in diffusion-derived metrics including mean diffusivity (MD) and fractional anisotropy (FA) and demonstrate improvements in test-retest results.
Methods
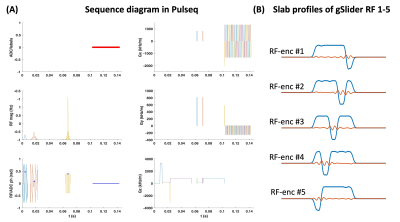
Pulse sequence programming and implementationThe sequence programming and the following data reconstruction were performed in MATLAB R2017a. Figure 1A presents the gSlider sequence diagram, while the corresponding slab profiles of five slab-encoding RFs are shown in Figure 1B. In Pulseq file, the sequence can be separated into blocks such as RF excitation, diffusion-sensitizing, refocusing, and EPI readout. With vendor-specific interpreters, the seq file which contains different blocks can be converted into hardware commands and executed on the scanners.
Data acquisition, reconstruction, and statistical analysis
The in-vivo experiments were carried out on a 3T scanner (MAGNETOM Prisma, Siemens Healthineers, Erlangen). Both vendor-based developed and Pulseq gSlider sequences (named Siemens-gSlider and Pulseq-gSlider) were used to obtain scan-rescan data on the same healthy volunteer to assess the test-retest reliability. The scanning parameters used for gSlider sequences were: FOV=220×220×130 mm3, in-plane image size: 220×220, 26 thin slabs (5-mm slab encoding), TR/TE = 3500/78 ms, and echo spacing=0.93 ms, in‐plane acceleration R=3, partial Fourier factor=6/8, 12 diffusion directions with b-value=1000, and 2 b-value=0 scans. FOV-matched B1+ maps were also acquired using Turbo‐FLASH8. GRAPPA was used to recover the missing k-space lines, after which the phase was removed from the complex data to obtain low-resolution real-valued images. The super-resolution reconstruction procedure from Liao et al9 was used to obtain the high-resolution data.
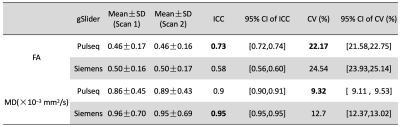
To calculate the test-retest variability, we calculated intraclass correlation coefficients (ICCs, two-way model, single measurement with absolute agreement) and coefficients of variation (CVs) for the FA and MD derived from Pulseq-gSlider and Siemens-gSlider in the white matter.
Results
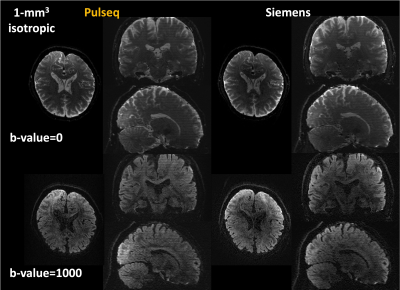
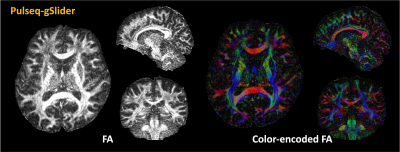
Figure 2 shows a comparison between isotropic-1mm dMRI data acquired from Pulseq-gSlider and Siemens-gSlider. Overall, Pulseq-gSlider presents detailed brain structures which are highly similar to the Siemens-gSlider data.FA maps and color-encoded FA maps calculated from dMRI acquired using Pulseq-gSlider are shown in Figure 3. With the ability of high-isotropic spatial resolution, fine-scale structures can be detected in all spatial views.
Figure 4 shows the evaluation of the test-retest reliability of the two gSlider sequences on the diffusion-derived metrics of white matter. Both sequences have excellent reliability in the MD measurement (ICC > 0.90) while presenting moderate reliability in the FA calculation (0.50<ICC<0.75)10. In FA, the ICC from Pulseq-gSlider is higher than Siemens-gSlider (0.73 vs 0.58). We also find that compared with Siemens-gSlider, the CVs from Pulseq-gSlider are slightly lower in both FA (22.17% vs 24.54%) and MD (9.32% vs 12.70%).
Discussion and Conclusion
Implementing and using advanced pulse sequences on different MRI scanners and vendors face significant challenges due to differences in software versions and manufacturers. Our vendor-independent Pulseq platform addresses this problem by describing the MRI sequence of events in a high-level format. Using the building blocks of Pulseq, we successfully implemented the high-isotropic resolution dMRI gSlider sequence. Comparable 1mm-isotropic dMRI are achieved using our implementation and the vendor-specific Siemens gSlider. Benefiting from the ultra-high isotropic spatial resolution, detailed structures can be observed from the diffusion tensor images in three views.As a prelude to acquiring harmonized MRI data across multiple sites/vendors, we also revealed the test-retest reliability of the Pulseq-gSlider sequence. Similar to Siemens-gSlider, our sequence demonstrated excellent reliability in MD values, whereas the reliability in FA measurements was moderate (potentially due to fewer gradient directions). As a preliminary study, we only tested Pulseq-gSlider on one scanner, future work will be focused on acquiring and carrying out similar experiments across various manufacturers.
Acknowledgements
This study is supported by NIH grants R01MH116173, R01MH125860, and R01EB032378.References
1. Chang H-C, Sundman M, Petit L, et al. Human brain diffusion tensor imaging at submillimeter isotropic resolution on a 3Tesla clinical MRI scanner. NeuroImage (Orlando, Fla) 2015;118:667–75.
2. Haldar JP, Liu Y, Liao C, Fan Q, Setsompop K. Fast submillimeter diffusion MRI using gSlider‐SMS and SNR‐enhancing joint reconstruction. Magn Reson Med 2020;84:762–76.
3. Engström M, Skare S. Diffusion-weighted 3D multislab echo planar imaging for high signal-to-noise ratio efficiency and isotropic image resolution. Magn Reson Med 2013;70:1507–14.
4. Setsompop K, Fan Q, Stockmann J, et al. High-resolution in vivo diffusion imaging of the human brain with generalized slice dithered enhanced resolution: Simultaneous multislice (gSlider-SMS). Magn Reson Med 2018;79:141–51.
5. Ramos-Llordén G, Ning L, Liao C, et al. High-fidelity, accelerated whole-brain submillimeter in vivo diffusion MRI using gSlider-spherical ridgelets (gSlider-SR). Magn Reson Med 2020;84:1781–95.
6. Layton KJ, Kroboth S, Jia F, et al. Pulseq: A rapid and hardware-independent pulse sequence prototyping framework. Magn Reson Med 2017;77:1544–52.
7. Nielsen JF, Noll DC. TOPPE: A framework for rapid prototyping of MR pulse sequences. Magn Reson Med 2018;79:3128–34.
8. Chung S, Kim D, Breton E, Axel L. Rapid B1+ mapping using a preconditioning RF pulse with turboFLASH readout. Magn Reson Med 2010;64:439–46.
9. Liao C, Stockmann J, Tian Q, et al. High-fidelity, high-isotropic-resolution diffusion imaging through gSlider acquisition with B1+ and T1 corrections and integrated ΔB0/Rx shim array. Magn Reson Med 2020;83:56–67.
10. Koo TK, Li MY. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J Chiropr Med 2016;15:155–63.
Figures