2016
Recovering slice location for unconventional acquisition plane with deep learning algorithm: cardiac magnetic resonance case
Habib Rebbah1 and Timothé Boutelier1
1Research & Innovation, Olea Medical, La Ciotat, France
1Research & Innovation, Olea Medical, La Ciotat, France
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Data Analysis
In cardiac MR field, the slice location along the long axis is a key parameter for the standardization proposed by the American heart association. We explore here the ability of a CNN to estimate it.Introduction
In order to propose a standardization of the heart studies, the American heart association (AHA) proposed a subdivision of the myocardium into segments1 mainly separated by their feed coronary artery. Such a subdivision is driven based on some anatomical structures, but also on slice locations along the long axis of the heart, divided into 3 groups: base, mid-cavity and apex. To reduce the manual interaction steps, we propose here to evaluate the ability of a convolutional neural network (CNN) to identify the slice location of cardiac magnetic resonance (CMR) images.Methods
The built algorithm takes as input a 2D CMR image and returns a label corresponding to the slice location. The three groups of slice locations ignore important variations of the heart shape and size. To reduce such an uncertainty, the number of classes is increased and depends on the sequence studied: 16 for CINE, 10 for perfusion, and 5 for MOLLI pre/post-injected and T2 prepared (T2p) sequences. We trained the CNN with images from those sequences to obtain five algorithms for each sequence. The figure 1 illustrates the approach.Each dataset was tacking from the HIBISCUS-STEMI database (NCT03070496) and incorporates a homogeneous distribution of slice positions from the base to the apex of the heart. Since all the sequences processed are time series sequences, the input 2D image was the first image for CINE and T2p, the last one for MOLLIs, and the image with highest contrast (evaluated as the standard deviation -sd- of the image) for perfusion. A ratio of 80/20% for training/evaluation subdivision of datasets was used for all the fitting processes. Table 1 details the methodology.
The exact position of the slices along the long axis are not perfectly identical for each patient of the database. This is due to anatomical variability and even variability due to the technician manipulations. Hence, we tested the 1-neighbor accuracy on validation dataset, which value is 1 if the estimated slice location is in the +/-1 interval around the true location and 0 outside.
The U-Net was built using Tensorflow framework and all the developments were done in Python.
Results and discussion
The figure 2 presents the results for all the sequences in boxplot and the table 2 summarizes them quantitatively. The accuracy obtained is low with a minimum for CINE sequence (0.04) and a maximum for perfusion sequence (0.45). However, the 1-neighbor accuracy is significatively higher since all the results are in the [0.84, 0.89] interval.Such performances reflect the uncertainty presents in the database used for training the CNN. We also explored a probabilistic CNN using the tensorflow-probability framework to manage this uncertainty, but the approach did not lead to increase the accuracy (not presented here), suggesting that the classical CNN converged correctly to the mean and was able to manage the uncertainty.
On the other hand, the obtained high value of 1-neighbor accuracy allows us to validate the algorithms. Indeed, the variability of the database used for the training is likely limited to a restricted interval around the true slice location. Additionally, for the purpose of the AHA delimitation, the slice locations are split into 3 levels. A fine division of the slice locations as tested here (up to 16 levels for the CINE sequence) is not required and the uncertainty of the obtained algorithms is admissible.
Conclusion
The proposed CNN approach successfully estimated the slice locations for CMR cases, despite the variability of the database.Acknowledgements
This work was supported by the RHU MARVELOUS (ANR-16-RHUS-0009). The authors particularly thank Pierre Croisille, Magalie Viallon, Nathan Mewton, Charles De Bourguignon, and Lorena Petrusca for data acquisition and management.References
1. Selvadurai, B. S. N. et al. Definition of Left Ventricular Segments for Cardiac Magnetic Resonance Imaging. JACC: Cardiovascular Imaging 11, 926–928 (2018).Figures
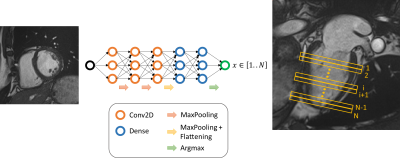
Figure 1 - CNN for slice location, illustration scheme.

Table 1 - CNN for slice location: databases details.
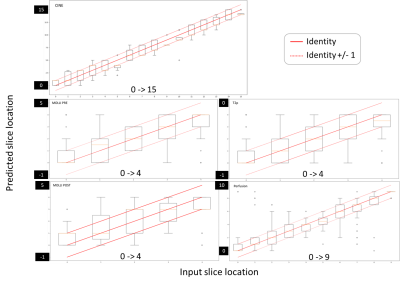
Figure 2 - CNN for slice location: results of evaluation datasets. Comparison of estimated slice position and inputs ones.
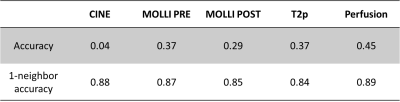
Table 2 - CNN for slice location: results in term of accuracy computed with evaluation datasets.
DOI: https://doi.org/10.58530/2023/2016