2012
Artifacts Map-guided Nonlocal Mean denoising for abdominal 4D-MR images acquired by 3D view-sharing sequence1Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, Hong Kong, 2Department of Biomedical Engineering, The Chinese University of Hong Kong, Hong Kong, Hong Kong, 3Peking University Cancer Hospital & Institute, Beijing, China, 4Department of Clinical Oncology, The University of Hong Kong, Hong Kong, Hong Kong, 5Department of Clinical Oncology, Queen Mary Hospital, Hong Kong, Hong Kong
Synopsis
Keywords: Data Processing, Data Processing, 4D-MRI
3D view-sharing sequences have been demonstrated its great promise for abdominal 4D-MRI due to its high temporal resolution and availability in clinical scanners. However, the sub-optimal sampling methods in these sequences under free-breathing condition deteriorates the image quality by ghost artifacts. Here, we propose a novel technique, Artifacts Map-guided Nonlocal Mean (AM-NLM), to suppress motion artifacts and to increase image quality of the 4D-MR images of liver cancer patients acquired by a 3D view-sharing sequence, TRICKS, on a 1.5T scanner.Purpose
This work aims to propose a novel technique, Artifacts Map-guided Nonlocal Mean (AM-NLM), that can suppress motion artifacts and enhance image quality of the 4D-MR images acquired by a clinically available 3D view-sharing sequence.Methods
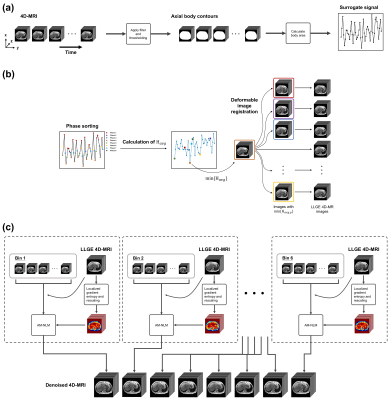
The proposed technique was validated on the 4D-MR images of 28 liver cancer patients acquired by a clinically available 4D view-sharing sequence, TRICKS, on a 1.5T scanner under free breathing condition. The scanning parameters were as follows: TR = 2.30 ms; TE = 0.816 ms; flip-angle = 10 o; matrix size = 256 × 256; voxel size = 1.37 × 1.37 × 5 mm3; acquisition time = 53 s; number of frames = 48; bandwidth = 488 Hz/pixel; temporal resolution = 1 s/volume.The overview of the 4D-MRI workflow involving the proposed AM-NLM is summarized in Fig.1. The surrogate signal was extracted by the axial body area (BA) method, 1-3 followed by sorting the images into 8 respiratory phases (Fig.1(a)). Subsequently, pixel-wise localized gradient entropy $$$\mathbf{H}(x,y,z)$$$ was calculated for every frames based on the sliding window technique and Hanning filtration. 4 The artifacts map was generated by normalization and histogram binning such that the maximum value was equal to the noise power $$$\sigma^2$$$, which was estimated in the background of the 4D-MR images. The frame with the global minimum average magnitude of $$$\mathbf{H}(x,y,z)$$$, namely $$$H_{avg}$$$, was sequentially registered to the frames with the minimum in each phase, and the resultant 4D-MR images were named as “LLGE 4D-MR images” (Fig.2(b)). The registration was performed using Elastix with B-spline interpolator. 5 In the final step (Fig.2(c)), the AM-NLM was implemented on the LLGE 4D-MR images. The proposed AM-NLM can be mathematically expressed as:
$$AMNLM(i)=\sum_{j,k\in I_p}w(i,j,k)v(j,k)$$ [1]
$$w(i,j,k)=\frac{1}{Z(i)}e^{-\frac{\widetilde{d}(i,j,k)}{[AM(i)]^2}}$$ [2]
, where $$$i$$$, $$$j$$$ and $$$k$$$ are the pixel index of the images $$$I_p$$$ in phase $$$p$$$, pixel index of the search window’s frame of reference with respect to the center $$$i$$$, and the index of the frame in phase $$$p$$$, respectively; $$$Z(i)$$$ is the normalization constant; $$$\widetilde{d}(i,j,k)$$$ is the approximated Euclidean distance; $$$AM$$$ is the artifacts map.
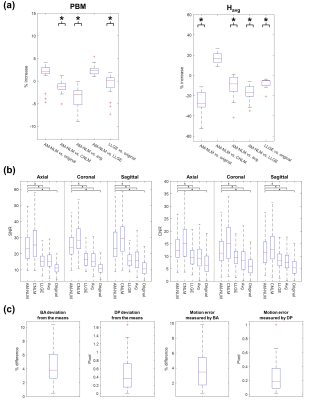
Image quality and motion accuracy of the 4D-MR images were evaluated quantitatively. Image quality was evaluated by signal-to-noise ratio (SNR), hepatic vein-to-liver parenchyma contrast-to-noise ratio (CNR), average localized gradient entropy ($$$H_{avg}$$$), and the perceptual blur metric (PBM). 6,7 The 4D-MR images generated by the AM-NLM approach were compared with that generated by other four approaches, namely conventional NLM, LLGE, simple averaging, and original 4D-MR images. Motion accuracy was evaluated by the deviation of BA (or diaphragm position, DP) of 4D-MR images from the mean BA (or DP) of the corresponding phase, and the deviation of BA (or DP) of 4D-MR images from the images with the minimum $$$H_{avg}$$$ of the corresponding phase (i.e. the pre-registered image).
Results
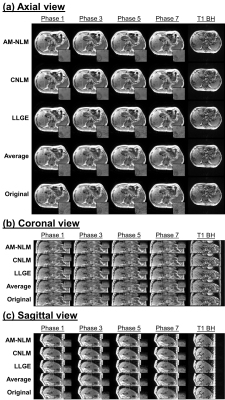
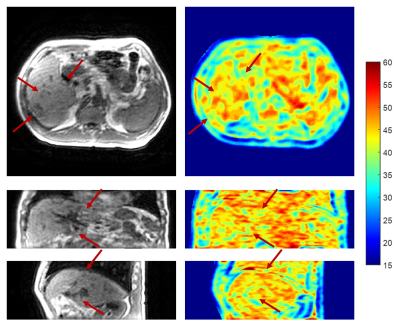
The 4D-MR images of a representative patient generated using different approaches are presented in Fig.2. The 4D-MR images generated by the proposed AM-NLM show significant improvement of image quality and retention of image sharpness in all three planes and all phases as compared to the other approaches. Despite better noise removal effect achieved by the CNLM approach, some fine anatomic details were over-smoothed. The great ability of edge preservation of the proposed AM-NLM was attributable to the guidance of the NLM by the artifacts map, as the associated smoothing parameters were effectively be suppressed at the locations with fine structures (Fig.3).Quantitative measurements of the 4D-MR images generated by different approaches are summarized in Fig.4. The AM-NLM 4D-MR images show significant enhancement in average SNR (CNR) over all phases and three planes as compared to the original 4D-MR images, with the magnitude of 25.9 ± 8.57 (13.0 ± 5.93) and 11.7 ± 3.86 (6.50 ± 3.27), respectively. The average deviation in DP and the registration errors for the 4D-MR images generated proposed AM-NLM approach were in a sub-pixel scale, indicating that a high motion accuracy was achieved by the AM-NLM approach.
Discussion
3D view-sharing sequences has been demonstrated its potential application in abdominal 4D-MRI due to its sub-second acquisition speed and clinical availability. 8,9 However, the sub-optimal use of the sequence leads to a great vulnerability to respiratory motion, and hence, a decrease in image quality. Here, we propose a novel technique, AM-NLM, that particularly aims to mitigate the motion artifacts of the TRICKS acquired 4D-MR images. Two major modifications were made as compared to the conventional NLM. 10,11 First, the smoothing parameter in the denominator of the exponential factor was replaced by the artifacts map. Second, the nonlocal searching patches were extended to the image frames in the same phase bin. This leaded to an increase of data availability for the averaging process. Moreover, the dynamic MR images defined the average motion of organs and tissues by means of deformable vector fields (DVFs) for the generation of LLGE 4D-MR images. Overall, the proposed approach allowed a fully exploitation of the spatial and temporal information of the acquired 4D-MR images for denoising.Conclusion
Both qualitative and quantitative results suggested that AM-NLM technique was capable of suppressing motion artifacts and improving image quality of the 4D-MR images acquired by a 3D view-sharing sequence.Acknowledgements
This work was partly supported by research funding from the GRF (15102118 and 15102219), and the HMRF (06173276).References
1. Cai J, Chang Z, Wang Z, Paul Segars W, Yin F-F. Four-dimensional magnetic resonance imaging (4D-MRI) using image-based respiratory surrogate: A feasibility study. Med Phys. 2011;38(12):6384-6394.
2. Liu Y, Yin F-F, Chang Z, et al. Investigation of sagittal image acquisition for 4D-MRI with body area as respiratory surrogate. Med Phys. 2014;41(10):101902.
3. Yang J, Cai J, Wang H, et al. Four-Dimensional Magnetic Resonance Imaging Using Axial Body Area as Respiratory Surrogate: Initial Patient Results. Int J Radiat Oncol Biol Phys. 2014;88(4):907-912.
4. Cheng JY, Alley MT, Cunningham CH, Vasanawala SS, Pauly JM, Lustig M. Nonrigid motion correction in 3D using autofocusing withlocalized linear translations. Magn Reson Med. 2012;68(6):1785-1797.
5. Klein S, Staring M, Murphy K, Viergever MA, Pluim J. elastix: A Toolbox for Intensity-Based Medical Image Registration. IEEE Trans Med Imaging. 2010;29(1):196-205.
6. Feuerlein S, Gupta RT, Boll DT, Merkle EM. Hepatocellular MR contrast agents: Enhancement characteristics of liver parenchyma and portal vein after administration of gadoxetic acid in comparison to gadobenate dimeglumine. Eur J Radiol. 2012;81(9):2037-2041.
7. Crete F, Dolmiere T, Ladret P, Nicolas M. The blur effect: perception and estimation with a new no-reference perceptual blur metric. Paper presented at: Human Vision and Electronic Imaging XII; 2007-02-15, 2007.
8. Plathow C, Schoebinger M, Herth F, Tuengerthal S, Meinzer H-P, Kauczor H-U. Estimation of Pulmonary Motion in Healthy Subjects and Patients with Intrathoracic Tumors Using 3D-Dynamic MRI: Initial Results. Korean Journal of Radiology. 2009;10(6):559.
9. Yuan J, Wong OL, Zhou Y, Chueng KY, Yu SK. A fast volumetric 4D-MRI with sub-second frame rate for abdominal motion monitoring and characterization in MRI-guided radiotherapy. Quantitative Imaging in Medicine and Surgery. 2019;9(7):1303-1314.
10. Buades A, Coll B, Morel J-M. A Non-Local Algorithm for Image Denoising. Paper presented at: 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR'05).
11. Tristán-Vega A, García-Pérez V, Aja-Fernández S, Westin C-F. Efficient and robust nonlocal means denoising of MR data based on salient features matching. Computer Methods and Programs in Biomedicine. 2012;105(2):131-144.
Figures