2007
Iterative Motion-Compensated Reconstruction with Convolutional Neural Network (iMoCo-Net) for Ultrashort Echo Time (UTE) Proton Lung MRI
Fei Tan1, Ke Wang2, Michael Lustig1,2, and Peder E. Z. Larson1,3
1UC Berkeley-UCSF Graduate Program in Bioengineering, University of California Berkeley and University of California, San Francisco, San Francisco, CA, United States, 2Electrical Engineering and Computer Sciences, University of California, Berkeley, Berkeley, CA, United States, 3Department of Radiology and Biomedical Imaging, University of California, San Francisco, San Francisco, CA, United States
1UC Berkeley-UCSF Graduate Program in Bioengineering, University of California Berkeley and University of California, San Francisco, San Francisco, CA, United States, 2Electrical Engineering and Computer Sciences, University of California, Berkeley, Berkeley, CA, United States, 3Department of Radiology and Biomedical Imaging, University of California, San Francisco, San Francisco, CA, United States
Synopsis
Keywords: Motion Correction, Motion Correction
This abstract explores the feasibility of machine learning-based motion-compensated reconstruction for free-breathing UTE lung MRI. Specifically, we used respiratory motion-resolved Non-uniform Fourier Transform (NuFFT) reconstruction as input, iterative motion-compensated (iMoCo) reconstruction as target, and a 2D U-Net convolutional neural network. Test results demonstrate a sharper diaphragm and a higher apparent SNR compared to the averaged input. In conclusion, iMoCo-Net accelerates the reconstruction of 3D radial UTE data substantially, shortening the required time from hours to minutes.Introduction
Proton pulmonary MRI is challenging due to the respiratory motion and low proton density. Breath-holding minimizes the motion but may not be tolerated by patients with severe diseases and the pediatric population, and also limits the spatial coverage and resolution due to the limited scan time. Free-breathing UTE sequence can produce high-quality proton lung MRI because it is more motion-robust compared to Cartesian sequences and exploits the fast-decaying signal. Motion-compensated reconstruction techniques (1–4) better manage respiratory motion for UTE acquisitions and are also more SNR efficient than respiratory-gated techniques. However, these methods are usually iteration-based optimization algorithms and can easily take hours to complete for a single dataset.Meanwhile, technologists prefer visualizing the reconstructed image during scanning to validate if a repeated scan is needed, and radiologists usually read the images within 1-2 hours from the scan. An accelerated motion-compensated reconstruction is needed. Luckily, machine-learning approaches can meet this demand. Among the established networks, U-Net (5) is well suited for image reconstruction and motion artifact removal tasks. While the training process is time-consuming, the inference for one subject only takes a minute. This abstract explores the feasibility of machine learning-based motion-compensated reconstruction for free-breathing UTE lung MRI.
Methods
Data AcquisitionTwenty subjects were retrospectively included in this study, ranging from healthy volunteers to pediatric and adult patients with lung diseases. Each subject underwent a 5-min free-breathing optimized radial golden angle UTE sequences (6) on a 3T clinical scanner (GE Healthcare, Milwaukee, WI) with a phased array coil. Key parameters were: resolution=1-1.5 mm, #spokes=80,000-100,000 spokes, TE/TR=0.07-0.1/2.7-3.8 ms.
Data Preparation
In order to form the network input, the raw data were separated into eight bins and reconstructed with Non-uniform Fourier Transform (NuFFT) reconstruction. The center of k-space was frequency filtered to track the respiratory motion for binning. Each motion state image corresponds to one input channel. We tested two sets of input, motion-resolved images reconstructed with all k-space spokes and the other mimicking an accelerated scan with only 50% of the spokes.
Iterative motion-compensated (iMoCo) reconstruction (7) results using all spokes were used as the target output for both cases. Depending on the number of coils, iMoCo reconstruction takes 1.5 to 6 hours to complete on our GPU server.
Center coronal slices that contain body structures were extracted as input and target images of the network. They were spatially normalized to matrix size 256x256, and the intensity was normalized to 0 to 1. The twenty data sets were separated into 16/2/2 patients for training, validation, and testing.
Network Structure
A 2D U-Net (5) was applied for the motion compensation task. Each encoding block includes a 3x3 padded convolutional layer, a rectified linear unit (ReLU) activation layer, and a batch normalization layer. A 2x2 max-pooling layer was applied between resolution scales. Similarly, convolution and ReLU layers were applied in the same scales for the decoding blocks, and a 2x2 up convolution was used between scales. The network was implemented in Tensorflow Keras with the mean absolute value error (L1) loss function and 50 epochs.
All processes, including data preparation and network training, were done on a GPU cluster, allocating one GPU (16 GB) core per task. The network structure details are summarized in Figure 1.
Results
Figures 2 and 3 demonstrate two example test set results from the neural network. The predicted image from all spokes preserves the fine vasculature structure of the lung. We also observe a sharper diaphragm and a higher apparent SNR compared to the averaged input. This indicates the network is successfully performing motion compensation and leveraging data from all motion states. As for the 50% spokes input, the prediction is significantly improved from the input but appears blurrier compared to the 100%-spoke results. The total reconstruction processing time for one subject is the time for NuFFT reconstruction (4 mins) and the time for inference (1 min for ~300 slices).The box plots of the structural similarity (SSIM), peak signal-to-noise ratio (pSNR), and mean squared error (MSE) metrics are shown in Figure 4. The mean structural similarity is 0.88±0.10 for 100% of spokes and 0.84±0.10 for 50% of spokes. The pSNR are 30.60±4.72 and 27.50±4.26, respectively, and the MSEs are 1.81e-3±3.07e-3 and 2.99e-3±3.60e-3.
Discussion
In this abstract, we used iMoCo reconstruction as target images, but it can be easily transferred to other motion-compensated reconstruction approaches and other organs that require motion compensation during image reconstruction, such as the heart and liver.While the primary purpose of this approach is to accelerate the reconstruction time for motion-compensated reconstructions for 3D data, results from 50% spokes suggest that neural network-based motion compensation can also facilitate shortened scan times. This improvement would benefit pediatric patients who fail to stay still for a 5 min scan or adult patients who experience involuntary motion.
Conclusion
This abstract demonstrated the feasibility of using a convolutional neural network for motion-compensated proton lung MRI reconstruction and motion artifact removal. It accelerates the reconstruction for 3D radial UTE data substantially, shortening the required time from hours to minutes. Furthermore, it also shows potential to shorten the scan time, thus facilitating the clinical application of proton pulmonary UTE MRI.Acknowledgements
No acknowledgement found.References
1. Feng L, Axel L, Chandarana H, Block KT, Sodickson DK, Otazo R. XD‐GRASP: Golden‐angle radial MRI with reconstruction of extra motion‐state dimensions using compressed sensing. Magn. Reson. Med. 2016;75:775–788 doi: 10.1002/mrm.25665.2. Jiang W, Ong F, Johnson KM, et al. Motion robust high resolution 3D free-breathing pulmonary MRI using dynamic 3D image self-navigator: Motion Robust High-Resolution 3D Pulmonary MRI. Magn. Reson. Med 2018;79:2954–2967 doi: 10.1002/mrm.26958.
3. Javed A, Ramasawmy R, O’Brien K, et al. Self‐gated 3D stack‐of‐spirals UTE pulmonary imaging at 0.55T. Magnetic Resonance in Med 2022;87:1784–1798 doi: 10.1002/mrm.29079.
4. Ding Z, Cheng Z, She H, Liu B, Yin Y, Du YP. Dynamic pulmonary MRI using motion‐state weighted motion‐compensation ( MOSTMOCO ) reconstruction with ultrashort TE : A structural and functional study. Magnetic Resonance in Med 2022;88:224–238 doi: 10.1002/mrm.29204.
5. Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. 2015.
6. Johnson KM, Fain SB, Schiebler ML, Nagle S. Optimized 3D ultrashort echo time pulmonary MRI. Magnetic Resonance in Medicine 2013;70:1241–1250 doi: 10.1002/mrm.24570.
7. Zhu X, Chan M, Lustig M, Johnson KM, Larson PEZ. Iterative motion‐compensation reconstruction ultra‐short TE (iMoCo UTE) for high‐resolution free‐breathing pulmonary MRI. Magn Reson Med 2020;83:1208–1221 doi: 10.1002/mrm.27998.
Figures
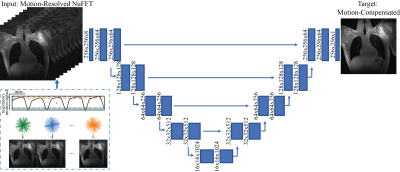
Figure 1. iMoCo-Net structure. The inputs are motion-resolved images reconstructed with NuFFT, binned based on k-space center data, and the target output are the iMoCo reconstructed image.
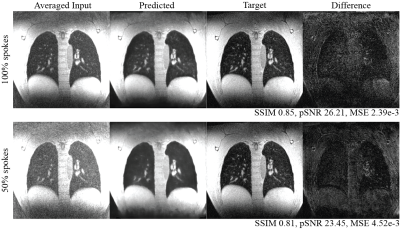
Figure 2. Example test set results with inputs generated using 100% and 50% of the raw data. In both cases, there is a clear apparent SNR improvement. The sharpness of the diaphragm is a good marker of motion compensation, and is particularly sharp when using 100% of the raw data.
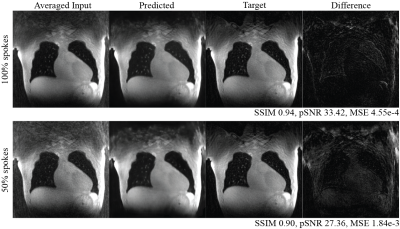
Figure 3. Additional example test set results with inputs generated using 100% and 50% of the raw data. Sharp diaphragm and small vessels were preserved in both cases.
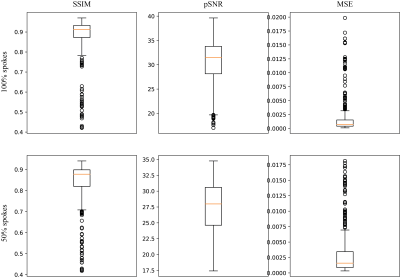
Figure 4. Structural similarity, peak signal-to-noise ratio, and mean squared error for 100% and 50% spoke motion-resolved NuFFT as network inputs.
Figure 5. Network loss functions showing clear convergence.
DOI: https://doi.org/10.58530/2023/2007