1967
Radiogenomic-based model identifies prognostic-related biological pathways in diffuse midline gliomas
Xiaorui Su1, Xibiao Yang1, Shuang Li1, Hanbing Shao1, Yukun Liu2, and Qiang Yue1
1Huaxi MR Research Center (HMRRC), Department of Radiology, West China Hospital of Sichuan University, Chengdu, China, 2West China School of Medicine &West China Hospital, Chengdu, China
1Huaxi MR Research Center (HMRRC), Department of Radiology, West China Hospital of Sichuan University, Chengdu, China, 2West China School of Medicine &West China Hospital, Chengdu, China
Synopsis
Keywords: Tumors, Radiomics
To explore biological interpretation of prognosis-related radiomic signatures in diffuse midline gliomas, forty-one cases with paired MRI and RNA sequencing were screened in the public database TCGA-TCIA. We extracted radiomics features from preoperative MRI and divided these patients into high- and low-risk groups. Differential and enrichment analysis showed that high-expression genes of the high-risk group were mainly activated and involved in biological processes such as axon extension, neurotransmitter signaling, and cell connection. Moreover, typical driver molecular changes known in DMG were observed. It suggested that biological interpretation of prognosis-related radiomic signatures could be achieved by integrating MRI and RNA dataINTRODUCTION
Diffuse midline glioma (DMG) is a type of malignant tumor that occurs in the midline of the brain, such as the pons of children and the thalamus of adolescents, with diffuse and invasive growth[1; 2]. The same molecular alterations in DMG have different prognostic outcomes, and the underlying biological mechanisms are not thoroughly understood. Therefore, more precise preoperative indicators are urgently needed to analyze tumor heterogeneity and correlate with patient prognosis, so as to achieve the purpose of precise treatment.METHODS
DMG samples with paired MRI and RNA sequencing were identified from the public data repository (TCGA-TCIA, The Cancer Imaging Archive. https://public.cancerimagingarchive.net/ncia/login.jsf, accessed April 2016). The radiomics feature were extracted form preoperative FLAIR and T1CE sequences. Main data processes in this study were as follows: using machine learning methods to select radiomic features related to DMG prognosis; building a prognosis prediction model based above radiomics features to generate high-risk group. Then, enrichment analysis was performed on the gene list generated by the differential analysis in the above steps. The key genes or pathways related to prognosis are identified by highly expressed genes of high-risk group in the differential analysis. Computation and statistical analysis are based on R language platform and Python packages. P values < 0.05 were considered statistically significant.RESULTS
There were 41 midline gliomas with preoperative MRI glioma were recruited in our study (mean age 59.90 years old, 18 male, 17 female, and 6 without gender information), including 27 GBM, 5 astrocytomas, 8 oligoastrocytomas and 1 oligodendroglioma. Total 1015 radiomics features were extracted from each subject, and after univariate regression, the remaining 102 features were subjected to LASSO-Cox regression analysis using machine learning and other methods. A prognostic model was constructed by screening the radiomics features related to the prognosis of DMG, and the RadRisk (RRS ) score of each sample was calculated based on the model. Finally, eight radiomics features are selected as prognostic-related radiomics features (see Figure 1 A):original_shape_Elongation,log_sigma_3_0_mm_3D_glcm_ClusterShade,wavelet_LHH_gldm_LargeDependenceHighGrayLevelEmphasis, wavelet_HLL_firstorder_Mean, wavelet_HLL_glcm_Imc2, wavelet_HLH_firstorder_Mean, wavelet_HHL_firstorder_Median, wavelet_HHL_glcm_Idmn.The survminer tool was used to determine the optimal RRS cutoff value of 8646.94, and patients were divided into high- and low- risk groups, including 31 and 10 subjects, there was a significant difference in OS between the two groups (P=0.018, see Figure 1B).After the differential analysis (Figure 2A), the results of GO enrichment analysis showed that the specific and highly expressed genes in the high-risk group were mainly activated and involved in biological processes such as axon extension, neurotransmitter signal transmission, and cell connection. The results of GSEA enrichment analysis also observed typical driver molecular changes known in DMG, such as the activation of biological pathways such as H3K27M me3 and PRC2 (see Figure 2B&C), which always mean poor prognosis.
Besides, in the Top50 gene panel (Figure 2A), we selected two genes with the most specific differential expression folds (Gene A and Gene B) to independently verify them in the public data of published articles. From the Bulk RNA-seq level, the expression levels of Gene A and Gene B in DIPG or GBM tumor types, the K27M mutant subgroup was significantly higher than the wild type; and Gene B was more specific in DIPG and significantly higher than the expression levels in GBM and normal brain tissue cortex (see Figure 3A&B).
DISCUSSION
Integrating correlative transcriptome data initially enables rational biological interpretation of prognosis-related radiomic features.In previous study, a group at Stanford University demonstrated[3] that neuroligin-3 produced by the electrical activity of neurons in the microenvironment can promote the formation of malignant gliomas. Subsequently, another study demonstrated[4] that malignant glioma cells can form excitatory synaptic connections with microenvironmental glutamatergic neurons to promote tumor proliferation and migration.The results of this study also suggest that neuronal activity may be responsible for the poor prognosis. In addition, we could observe significant suppression of immune activation-related genes in the high-risk group, suggesting that DMG may lead to poor prognostic outcomes by creating an immunosuppressive environment.
Our study demonstrated that the power potential of radiomics features in improving the precise diagnosis of DMG.
CONCLUSION
Integrating RNA-sequencing and MRI radiomics features could enable plausible biological interpretation of prognosis-related radiomics features in diffuse midline gliomas.Acknowledgements
The results shown here are in whole based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.References
- Price G, Bouras A, Hambardzumyan D, Hadjipanayis CG (2021) Current knowledge on the immune microenvironment and emerging immunotherapies in diffuse midline glioma. EBioMedicine 69:1034532
- Arakaki AKS, Szulzewsky F, Gilbert MR, Gujral TS, Holland EC (2021) Utilizing preclinical models to develop targeted therapies for rare central nervous system cancers. Neuro Oncol 23:S4-s153
- Venkatesh HS, Johung TB, Caretti V et al (2015) Neuronal Activity Promotes Glioma Growth through Neuroligin-3 Secretion. Cell 161:803-8164
- Venkataramani V, Tanev DI, Strahle C et al (2019) Glutamatergic synaptic input to glioma cells drives brain tumour progression. Nature 573:532-538
Figures
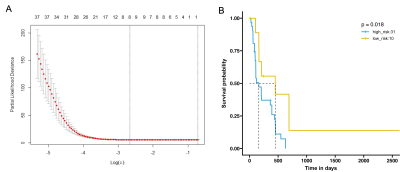
Figure 1. A: Final 8 radiomics features associated with prognosis; B: Survival analysis of high- and low-risk groups with optimal RRS thresholds

Figure 2. Panel formulation of Top50 mutation-related genes (A) and enrichment of some related pathways (B). The activation of biological pathways, H3K27M me3 and PRC2 (C). Gene A and Gene B are genes that are significantly up-regulated in the high-risk group
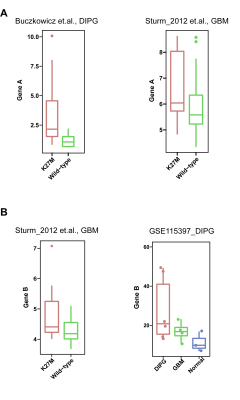
Figure 3. Public database transcription and expression of key genes; A: Gene A in the K27M mutant in DIPG and GBM has a significantly higher transcriptional expression level than in the wild type; B: Gene B in the K27M mutant of DIPG has a significantly higher transcriptional expression level than in the wildtype and more specific in DIPG.(data from ①Buczkowicz P, et al. Nat Genet. 2014; 46(5):451-456;②Sturm D, et al. Cancer Cell. 2012; 22(4):425-437;③GSE115397_DIPG:Lin GL, et al. Acta Neuropathol Commun. 2018; 6(1):51)
DOI: https://doi.org/10.58530/2023/1967