1966
MRI-Based Radiomics Approach with Deep Learning for Distinguishing IDH-Mutant from IDH Wild-type Grade-4 Astrocytomas1Department of Neurology and Neurosurgery, Faculty of Medicine, McGill University, Montréal, QC, Canada, 2Translational Neuroimaging Laboratory, The McGill University Research Centre for Studies in Aging, Douglas Hospital,McGill University, Montreal, QC, Canada, 3Electrical and Computer Engineering, Kharazmi University, Tehran, Iran (Islamic Republic of), 4Division of Nuclear Medicine and Molecular Imaging, Geneva University Hospital, Geneva, Switzerland, 5Rajaie Cardiovascular Medical and Research Center, Iran University of Medical Science, Tehran, Iran (Islamic Republic of), 6Radiology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, United States, 7Hematology-Oncology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, United States, 8Clinical Pathology and Laboratory Medicine, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, United States, 9Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, United States
Synopsis
Keywords: Tumors, Radiomics
Patients (n=57) with isocitrate dehydrogenase (IDH) mutant grade-4 astrocytomas and IDH wild-type glioblastomas underwent anatomical imaging (post-contrast T1 and T2-FLAIR) on 3T magnet. Neoplasms were segmented into 5 ROIs and 105 radiomics features were extracted from each ROI. Features were subsequently selected using various algorithms. Patients were divided into two groups (50%-training and 50%-testing). A GAN-based deep learning algorithm was used to generate 1000 synthesized data-sets and four distinct deep-learning modules were implemented. The best model for differentiating two genotypes of neoplasms was obtained from core tumor regions by using K-best feature selection and Ensembled algorithm with high diagnostic performance.Introduction
Isocitrate dehydrogenase (IDH) gene mutation occurs in 70% of WHO grade 2/3 gliomas and 10% of grade-4 astrocytomas (1). Despite the low occurrence rates of IDH mutation in grade-4 astrocytomas; it is crucial to develop imaging biomarkers to distinguish their IDH profiles for prognostication of these patients and formulating effective treatment plans. Radiomics and machine/deep learning methods have shown a great potential in making correct diagnosis, prognosis, and determining molecular signatures, assessing treatment response, and predicting survival outcomes in these patients (2, 3). Some other studies have also reported promising findings in identifying IDH mutant grade-4 astrocytomas using conventional neuroimaging-based radiomic classification models with variable accuracies. However, these studies were limited by the extraction of a sparse number of radiomic features (n=31) (4) or by the inclusion of a small sample size of IDH mutant grade-4 astrocytomas (n=7) (5).Purpose
The purpose of the current study was to build a predictive model using artificial intelligence and quantitative radiomics data extracted from conventional MR images and to investigate its potential in discriminating IDH-mutant grade-4 astrocytomas from IDH wild-type glioblastomas (GBMs).Methods
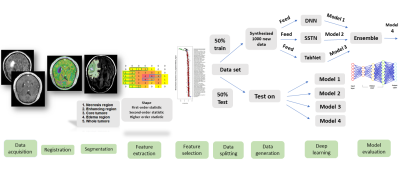
A cohort of 57 treatment naïve patients with IDH-mutant grade-4 astrocytomas (n=23) and IDH wild-type GBMs (n=34) underwent anatomical imaging on a 3T MR system with standard parameters. Post-contrast T1-weighted and T2-FLAIR images were resampled, and co-registered. Regions of interest (ROIs) were drawn manually from contrast-enhancing/solid regions, central necrotic region, and tumor core (solid + necrotic regions), on post-contrast T1 weighted images. Additionally, T2-FLAIR images were used to segment the peri-tumor signal abnormality and whole tumor (solid + necrotic tumor + peritumoral T2-FLAIR hyperintensity) in each case. Altogether, 105 first, second-, and higher-order statistics features along with shape-based features were extracted from each ROI using an open-source image biomarker standardization initiative compliant PyRadiomics package in python (6). Overall, 525 radiomics features were extracted from each image from 5 ROIs per patient. Various feature selection algorithms were employed to select radiomic features, including minimum redundancy maximum relevance (MRMR), K-best, and recursive feature elimination (RFE). Data were randomly divided into training (50%) and testing (50%) sets. To address the issue of the small number of real data sets for implementing deep learning algorithms, a GAN-based deep learning data synthesizer (CTGAN) algorithm (7) was implemented to generate 1000 synthesized data sets from the training set’s selected radiomic features. Four different deep learning modules were implemented in the current study. The first method was Deep Neural Net (DNN) with 4 dense layers and 2 dropout layers (Keras library in python platform). Loss function, binary-cross entropy, and optimizer Adadelta were used for data learning. The second method was TabNet, a deep tabular data learning architecture that encompassed 4 network-layer: 2 shared across all decision steps and 2 decision step-dependent. The third method was self-supervised TabNet (SSTN) which created a pre-trained model of our data set using self-supervised modules. The fourth method was the Ensemble model, which was a combination of these three models. The study schema is shown in Figure 1. To investigate the diagnostic performance of our classification models, receiver operative characteristic curve (ROC) analyses were performed and area under the ROC curve (AUC), accuracy (ACC), sensitivity (SEN), and specificity (SPE) were determined.Results
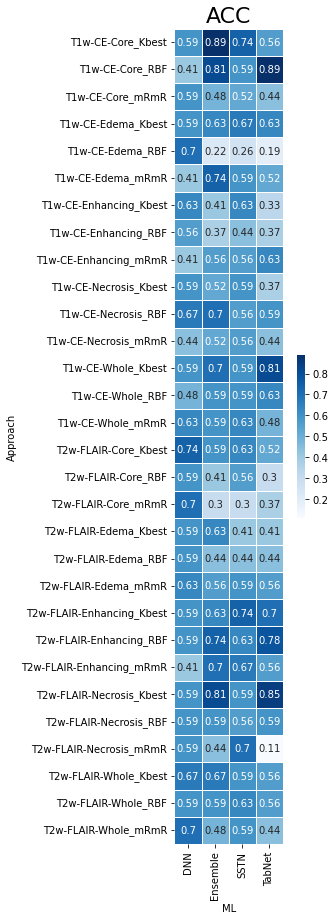
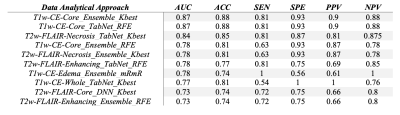
As presented in Table 1, the best model for differentiating IDH-mutant grade=4 astrocytomas from IDH wild-type GBMs was obtained from core tumor regions of neoplasms as visible on post-contrast-T1 co-registered images by using the K-best feature selection and Ensemble algorithm with high diagnostic performance (AUC= 0.87, ACC= 0.88, SEN= 0.81, SPE= 0.93). Similar findings were obtained from core tumor regions of T1-post contrast co-registered images using the RFE feature selection and TabNet algorithm. Heatmaps of predictive accuracy (ACC), predictive power (AUC), and sensitivity (SEN) for discriminating two genotypes of neoplasms utilizing a variety of feature selection and machine learning algorithms applied to distinct neoplastic subregions are shown in Figures 2-4 respectively.Discussion
This study developed a predictive model based upon multi-deep learning approach from different sub-regions of neoplasms as visible on post-contrast T1 images and T2-FLAIR in distinguishing IDH-mutant grade-4 astrocytomas from IDH-wild-type GBMs. Our work is an extension of previous studies as we used GAN based algorithm to increase our sample size and a large of machine learning classifiers (n=18) to build a reliable prediction model in distinguishing two genotypes of neoplasms.Conclusion
Conventional neuroimaging-derived radiomics and deep learning methods may be helpful in differentiating IDH-mutant grade-4 astrocytomas from IDH wild-type GBMs with high diagnostic performance.Acknowledgements
No acknowledgement found.References
1. Vigneswaran K, Neill S, Hadjipanayis CG. Beyond the World Health Organization grading of infiltrating gliomas: advances in the molecular genetics of glioma classification. Annals of translational medicine. 2015;3(7).
2. Hosseini SA, Hajianfar G, Shiri I, Zaidi H, editors. Lung Cancer Recurrence Prediction Using Radiomics Features of PET Tumor Sub-Volumes and Multi-Machine Learning Algorithms. 2021 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC); 2021: IEEE.
3. Hosseini SA, Hajianfar G, Shiri I, Zaidi H, editors. Lymphovascular Invasion Prediction in Lung Cancer Using Multi-Segmentation PET Radiomics and Multi-Machine Learning Algorithms. 2021 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC); 2021: IEEE.
4. Lee MH, Kim J, Kim S-T, Shin H-M, You H-J, Choi JW, et al. Prediction of IDH1 mutation status in glioblastoma using machine learning technique based on quantitative radiomic data. World neurosurgery. 2019;125:e688-e96.
5. Hsieh KL-C, Chen C-Y, Lo C-M. Radiomic model for predicting mutations in the isocitrate dehydrogenase gene in glioblastomas. Oncotarget. 2017;8(28):45888.
6. Van Griethuysen JJ, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, et al. Computational radiomics system to decode the radiographic phenotype. Cancer research. 2017;77(21):e104-e7.
7. Gupta A, Bhatt D, Pandey A. Transitioning from Real to Synthetic data: Quantifying the bias in model. arXiv preprint arXiv:210504144. 2021.
Figures