1895
GAMER-MRI and modified Layer-wise Relevance Propagation identify on quantitative MRI regions sensitive to clinical disability in MS patients1Translational Imaging in Neurology (ThINk) Basel, Department of Medicine and Biomedical Engineering, University Hospital Basel and University of Basel, Basel, Switzerland, 2Department of Neurology, University Hospital Basel, Basel, Switzerland, 3Research Center for Clinical Neuroimmunology and Neuroscience Basel (RC2NB), University Hospital Basel and University of Basel, Basel, Switzerland, 4AI for Clinical Analytics, Covera Health, New York, NY, United States, 5Division of Radiological Physics, Department of Radiology, University Hospital Basel, Basel, Switzerland, 6Center for medical Image Analysis & Navigation, Department of Biomedical Engineering, University of Basel, Allschwil, Switzerland, 7Neuroradiology Department, Inselspital, Bern, Switzerland
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Multiple Sclerosis
The decision process of artificial intelligence is elusive. We proposed a new method that by combining an attention-based convolutional neural network (GAMER-MRI) with the modified Layer-wise Relevance Propagation could reveal relevant regions on quantitative imaging maps in differentiating multiple sclerosis patients with mild-moderate and severe disabilities. The assessment of the relevant regions included the impact of inverting values within the regions and the heatmap on the MNI152 template. Our results show good network performance and identify brain regions relevant to the corticospinal tract. The proposed method might be useful to further explore patterns of brain microstructural alterations associated with disability.Introduction
Quantitative MRI provides microstructural measures of brain tissue property relating to its main components (e.g. myelin – myelin water fraction (MWF) and quantitative T1 (qT1); axon, qT1 and Neurite density Index (NDI))1. Those quantitative MR measures (qMRs) are especially suitable to investigate multiple sclerosis (MS) pathology because this latter typically affects the integrity of myelin and axons2. Using qMRs, we successfully developed an attention-based convolutional neural network – GAMER-MRI – to obtain attention weights as quantitative proxies for the importance of qMRs in MS applications3,4. However, this approach suffered a common issue of deep neural networks that the deep layer structure and nonlinear activations hinder the understanding of the decision process5. The Layer-wise Relevance Propagation (LRP)5 alleviates this issue by using various rules to redistribute the output based on the contributions of neurons in the network, hereby providing with relevance maps. To find out which brain regions on qMR maps considered by GAMER-MRI determine the classification of MS patients with mild vs severe disability, we proposed a new approach combining GAMER-MRI and modified LRP.Methods
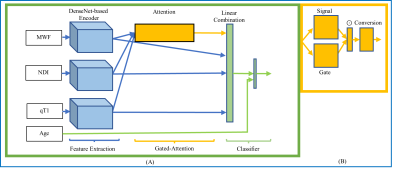
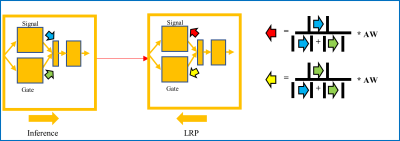
We enrolled 166 MS patients (100 relapsing-remitting and 66 progressive, 99 females, age range=45.9±14.3). Their disability was measured using the Expanded Disability Status Scale (EDSS, median=2.5). Patients underwent MRI on a 3T whole-body MR system (Siemens MAGNETOM Prisma). Sixty-nine of the patients underwent 2-year follow-up MRI acquisitions. MRI data were reconstructed into three qMRs3: MWF of 0.94x0.94x5 mm3, NDI of 1.8x1.8x1.8 mm3, and qT1 of 1x1x1 mm3. Non-brain tissue was removed by FreeSurfer6. qMRs were scaled between 0 and 1 according to the feasible value ranges within brain: MWF ≤30% and qT1 ≤2500 ms. qMRs were co-registered to the NDI reference space using FSL7. Forty patients with EDSS ≥5 (severe motor impairment) were assigned to the group with severe disability. The rest of the patients formed the mild-moderate disability group. An independent test dataset (16 in the mild disability group and 5 in the severe group) and a dataset for the stratified three-fold cross-validation were used considering the size of the severe group. GAMER-MRI included feature extraction, gated-attention mechanism, and classification (Fig. 1). The feature extraction was based on DenseNet8 with 16 filters at the first 3D convolutional layer, four dense blocks, two dense layers per dense block, and the growth rate equal to four. The hidden feature vectors from the feature extraction had 32 elements. The gated-attention mechanism consisted of the gate and signal branches, where the numbers of neurons in the layers were 16. Patient age was divided by 100 and concatenated to the combined hidden feature vector after the gated-attention mechanism. The classifier was a one-neuron fully connected layer. The binary cross entropy loss was weighted by $$$weight=2-\frac{|EDSS-5|}{5}$$$ so that the heterogeneity within groups could be better reflected during training. The evaluation metric was the area under the receiver operating characteristic curve (AUC). To alleviate overfitting, data augmentation included random flipping, random 90-degree rotation, random Gaussian noise of zero mean and standard deviation equal to 0.1, and random affine transformation with maximum rotation ±30 degrees and ±20% scaling. In addition, AdamW9 (a regularized optimizer) with a learning rate equal to 0.00005 was used. The weighted sampler was implemented to tackle the group imbalance. The modified LRP included redistribution based on attention weights, absolute contributions of the signal and gate branches (Fig. 2), and the combination of the relevance maps and the qMR maps. The relevant regions at different thresholds for the relevance values were obtained. To assess if relevant regions were relevant, the qMR values within the regions were inverted ($$$qMR=1-qMR$$$) and the subsequent reduction of the AUC was the measure of the importance of the regions. Regions obtained by the original LRP and other explainability methods (Saliency10 and Integrated Gradients11) underwent the same procedure for comparison. To explore the potential group effect, the relevant regions of patients were transformed to the MNI152 template12Results and Discussion
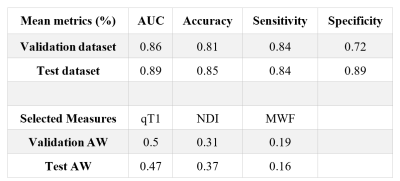
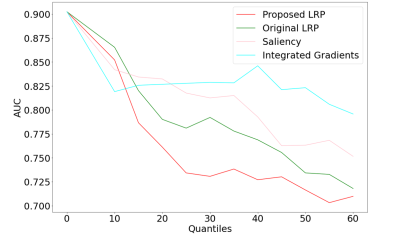
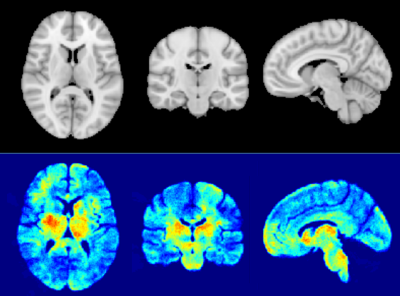
In Table 1, we report the average performance on the cross-validation folds and the test dataset. The performance indicated GAMER-MRI learned useful representation for further interpretation. According to the obtained attention weights, qT1 was the measure that best discriminated clinical severity in our cohort of MS patients, followed by NDI. This might be due to qT1 providing a comprehensive representation of the damaged microenvironment or more details revealed by the higher resolution and white-grey matter contrast. The reduction of the AUC in Fig. 3 shows that the proposed method revealed the most relevant regions. This resonates with our previous findings that the attention weights are importance proxies of qMRs. Identified regions on the MNI152 template in Fig. 4 include the left caudate, the left thalamus, the putamens, and the internal capsules. Those are structures that are either part of the corticospinal tract or regions involved in motor circuits, which are strongly related to EDSS and disability13.Conclusions
In summary, our work showed that the proposed combined approach could classify patients having a severe motor impairment and identify relevant regions on qMR maps. Future work will aim at (i) investigating the pathological meaning of the relevant regions; (ii) integrating other qMRs such as quantitative susceptibility mapping and magnetization transfer saturation.Acknowledgements
This project is supported by Swiss National Funds PZ00P3_154508, PZ00P3_131914 and PP00P3_176984 and we thank all the patients for their participation.References
1. Granziera C, Wuerfel J, Barkhof F, et al. Quantitative magnetic resonance imaging towards clinical application in multiple sclerosis. Brain. 2021;144(5):1296-1311. doi:10.1093/brain/awab029
2. Reich DS, Lucchinetti CF, Calabresi PA. Multiple Sclerosis. Longo DL, ed. N Engl J Med. 2018;378(2):169-180. doi:10.1056/NEJMra1401483
3. Lu PJ, Yoo Y, Rahmanzadeh R, et al. GAMER MRI: Gated-attention mechanism ranking of multi-contrast MRI in brain pathology. NeuroImage Clin. 2021;29:102522. doi:10.1016/j.nicl.2020.102522
4. Lu PJ, Barakovic M, Weigel M, et al. GAMER-MRI in Multiple Sclerosis Identifies the Diffusion-Based Microstructural Measures That Are Most Sensitive to Focal Damage: A Deep-Learning-Based Analysis and Clinico-Biological Validation. Front Neurosci. 2021;15. doi:10.3389/fnins.2021.647535
5. Samek W, Montavon G, Lapuschkin S, Anders CJ, Muller KR. Explaining Deep Neural Networks and Beyond: A Review of Methods and Applications. Proc IEEE. 2021;109(3):247-278. doi:10.1109/JPROC.2021.3060483
6. Fischl B. FreeSurfer. Neuroimage. 2012;62(2):774-781. doi:10.1016/j.neuroimage.2012.01.021
7. Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM. Review FSL. Neuroimage. Published online 2012. doi:10.1016/j.neuroimage.2011.09.015
8. Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. In: Proceedings - 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017. ; 2017. doi:10.1109/CVPR.2017.243
9. Loshchilov I, Hutter F. Decoupled weight decay regularization. In: 7th International Conference on Learning Representations, ICLR 2019. ; 2019.
10. Simonyan K, Vedaldi A, Zisserman A. Deep inside convolutional networks: Visualising image classification models and saliency maps. 2nd Int Conf Learn Represent ICLR 2014 - Work Track Proc. Published online 2014:1-8.
11. Sundararajan M, Taly A, Yan Q. Axiomatic attribution for deep networks. 34th Int Conf Mach Learn ICML 2017. 2017;7:5109-5118.
12. Grabner G, Janke AL, Budge MM, Smith D, Pruessner J, Collins DL. Symmetric Atlasing and Model Based Segmentation: An Application to the Hippocampus in Older Adults. In: ; 2006:58-66. doi:10.1007/11866763_8
13. Tovar-Moll F, Evangelou IE, Chiu AW, et al. Diffuse and Focal Corticospinal Tract Disease and Its Impact on Patient Disability in Multiple Sclerosis. J Neuroimaging. 2015;25(2):200-206. doi:10.1111/jon.12171
Figures