1891
RiskForm: A novel risk formulation to improve progressive disease outcome prediction
Haresh Rengaraj Rajamohan1, Kyunghyun Cho1, Richard Kijowski2, and Cem M. Deniz2
1New York University, NEW YORK, NY, United States, 2NYU Langone Health, NEW YORK, NY, United States
1New York University, NEW YORK, NY, United States, 2NYU Langone Health, NEW YORK, NY, United States
Synopsis
Keywords: Osteoarthritis, Machine Learning/Artificial Intelligence, Deep Learning
We propose a novel risk constraint to improve the performance of deep learning models on progressive disorders. On the Osteoarthritis Initiative (OAI) dataset, the proposed approach outperforms a baseline model trained with the standard cross-entropy loss on predicting total knee replacement (TKR) within 3 different time horizons- 1 year, 2 years and 4 years of the MRI date. It further generalizes better to the external Multicenter Osteoarthritis Study dataset.INTRODUCTION
Knee Osteoarthritis (OA) is the most common form of arthritis and a major cause of physical disability1. Due to the lack of an effective treatment for OA, an early diagnosis with behavioral interventions remains the best viable option to keep OA in check 2. To facilitate this, it's important to accurately predict if a patient’s knee will progress to undergo TKR eventually. In recent years, deep learning (DL) methods have shown great potential in MR image analysis on a variety of diseases including OA3-9. In this study, we propose adding a risk constraint specific to knee OA that ensures the risk of TKR for a knee, should either increase or remain the same over time. This assumption comes from the fact that OA is a progressive disease10. DL models were trained to predict TKR within 1 year, 2 years, and 4 years using two images of each patient's knee recorded at least 12 months apart and during training, this constraint was enforced between the two images. The modified risk formulation approach outperforms a standard baseline model on the Osteoarthritis Initiative (OAI)11 dataset and further generalizes better to the external Multicenter Osteoarthritis Study (MOST)12 dataset.METHODS
In this study, Coronal intermediate-weighted turbo spin echo (COR IW TSE) images from the OAI dataset were used for training and evaluation, and coronal short-tau inversion recovery (COR STIR) images from the MOST dataset were used for evaluation alone. In pre-processing, images were resized to 352x352x35 and standard normalized. Case-control matched cohorts of 708 knees from the OAI dataset and 756 knees from the MOST dataset were selected (Figure 1). The matching was performed based on age, BMI, ethnicity, and sex. For each patient's knee, two MR images were utilized- 1st image at the baseline visit and 2nd image is the last available MRI in the database (for TKR-positive knees, acquired before TKR). This scheme enabled us to apply our constraint and track the knee's OA progression. The knees that had only one image in the dataset were also used without the application of the risk constraint during training.Modified Risk Formulation
In the baseline approach, we have MR images of a knee from time-step-1 ($$$x_1$$$) and time-step-2 ($$$x_2$$$), CNN model ($$$f$$$), and sigmoid function ($$$\sigma$$$). The risk of TKR in $$$t$$$ years for the two images is,$$p(y^{1}_t=1\,|x_1)=\hat{y}^{1}_t=\sigma(f(x_1))$$
$$p(y^{2}_t=1\,|x_2)=\hat{y}^{2}_t=\sigma(f(x_2))$$
where $$$y^{1}_t,y^{2}_t\in{0,1}$$$ indicating TKR status in $$$t$$$ years for $$$x_1$$$ and $$$x_2$$$ respectively, $$$t\in{1,2,4}$$$, $$$f(x_1),f(x_2)\in R$$$ , $$$\sigma(f(x1)),\sigma(f(x2))\in(0,1)$$$ and $$$\hat{y}^{1}_t,\hat{y}^{2}_t$$$ are the TKR risk predictions. Our goal is to enforce the constraint $$$\hat{y}^{1}_t\leq\hat{y}^{2}_t$$$. To achieve this, the TKR risk computation for $$$x_1,x_2$$$ is modified as follows (RiskForm1),
$$p(y^{1}_t=1\,|x_1)=\hat{y}^{1}_t=\sigma(f(x_1))$$
$$p(y^{2}_t=1\,|x_2,x_1)=\hat{y}^{2}_t=1-p(y^{1}_t=0\,|x_1)*(1-\sigma(f(x_2))$$
Since $$$(1-\sigma(f(x_2))\in(0,1)$$$, $$ p(y^{1}_t=0\,|x_1)*(1-\sigma(f(x_2))\leq p(y^{1}_t=0\,|x_1) $$
$$\implies\,1- p(y^{1}_t=0\,|x_1)*(1-\sigma(f(x_2))\geq 1-p(y^{1}_t=0\,|x_1)$$
$$\implies\,p(y^{2}_t=1\,|x_2,x_1)\geq p(y^{1}_t=1\,|x_1)\implies\hat{y}^{1}_t\leq\hat{y}^{2}_t$$
Here, we are explicitly conditioning the TKR likelihood of $$$x_2$$$ on $$$x_1$$$. We further introduce an additional formulation RiskForm2 that utilizes two models $$$f,g$$$ and the TKR risks are
$$p(y^{1}_t=1\,|x_1)=\hat{y}^{1}_t=\sigma(f(x_1))$$
$$p(y^{2}_t=1\,|x_2,x_1)= \hat{y}^{2}_t=1- p(y^{1}_t=0\,|x_1)*(\sigma(g(x_2))$$. Here $$$x_1$$$ and $$$x_2$$$ are processed by two separate models. The training loss for the models is,
$$Loss=BCE(y^{1}_t,\hat{y}^{1}_t)+BCE(y^{2}_t,\hat{y}^{2}_t) $$
where BCE is the binary cross-entropy loss. In the case of a knee having only a single image, the TKR risk is $$$\hat{y}^{1}_t$$$ and $$$Loss=BCE(y^{1}_t,\hat{y}^{1}_t)$$$ is applied.
Models and Training
The OAI cohort was split in a subject-stratified manner into seven folds. Nested cross-validation (CV) with 3D-Resnet34 architecture13 and Adam optimizer14 (lr=2e-4) were used in training. In seven-fold nested CV, with each fold as a test set, six models were trained and they were saved based on the best overall AUC on their respective validation sets. During evaluation, on OAI set, each fold was evaluated by an ensemble of its six models and the metrics were computed over all the available knee images. On the MOST set, all 42 models were ensembled to compute the predictions.Results
Table 2 shows that all models perform well on the OAI set, achieving ~0.8 area under the ROC curve (AUROC) on all the tasks. There was a significant drop in the MOST performance, probably due to the differences between MR scanner and protocol (Figure 2). The modified risk formulation approach outperforms the baseline method on the OAI set and further generalizes better to the external MOST set. The proposed RiskForm2 approach, which utilizes two separate models to predict risk on the two MR images of a knee, achieves the best performance in terms of AUROC and AUPRC in most of the experimental settings. Figure 3 shows the AUROC achieved by the models on the OAI TKR positive (< 9year) subset. The superior performance of RiskForm methods might be indicative of their ability to better recognize the subtle differences in TKR-positive knees.DISCUSSION
The models trained using our proposed approach outperformed the conventional baseline. Their superior generalization performance on an external dataset suggests that incorporating the risk constraint makes these models more robust to changes in data distribution. An interesting future work would be to come up with a formulation that can effectively condition on more than a single past imageCONCLUSION
The proposed risk formulation will benefit the research community and could be applied to other progressive disorders to build better, more generalizable DL models for progressive outcomes' predictions.Acknowledgements
This work was supported in part by NIH grant R01 AR074453, and was performed under the rubric of the Center for Advanced Imaging Innovation and Research (CAI2R, www.cai2r.net), a NIBIB Biomedical Technology Resource Center (NIH P41 EB017183).References
- Cross, Marita, et al. "The global burden of hip and knee osteoarthritis: estimates from the global burden of disease 2010 study." Annals of the rheumatic diseases 73.7 (2014): 1323-1330.
- Karsdal, M. A., et al. "Disease-modifying treatments for osteoarthritis (DMOADs) of the knee and hip: lessons learned from failures and opportunities for the future." Osteoarthritis and cartilage 24.12 (2016): 2013-2021.
- Tolpadi, Aniket A., et al. "Deep learning predicts total knee replacement from magnetic resonance images." Scientific reports 10.1 (2020): 1-12.
- Panfilov, Egor, et al. "Deep learning‐based segmentation of knee MRI for fully automatic subregional morphological assessment of cartilage tissues: Data from the Osteoarthritis Initiative." Journal of Orthopaedic Research® 40.5 (2022): 1113-1124.
- Schiratti, Jean-Baptiste, et al. "A deep learning method for predicting knee osteoarthritis radiographic progression from MRI." Arthritis research & therapy 23.1 (2021): 1-10.
- Pedoia, Valentina, et al. "Diagnosing osteoarthritis from T2 maps using deep learning: an analysis of the entire Osteoarthritis Initiative baseline cohort." Osteoarthritis and cartilage 27.7 (2019): 1002-1010.
- Akkus, Zeynettin, et al. "Deep learning for brain MRI segmentation: state of the art and future directions." Journal of digital imaging 30.4 (2017): 449-459.
- Noor, Manan Binth Taj, et al. "Detecting neurodegenerative disease from MRI: a brief review on a deep learning perspective." International conference on brain informatics. Springer, Cham, 2019
- Herent, P., et al. "Detection and characterization of MRI breast lesions using deep learning." Diagnostic and interventional imaging 100.4 (2019): 219-225.
- Castañeda, Santos, et al. "Osteoarthritis: a progressive disease with changing phenotypes." Rheumatology 53.1 (2014): 1-3.
- Peterfy, C.G., M.D., Ph.D, Schneider E, Ph.D, Nevitt M, Ph.D. The osteoarthritis initiative: Report on the design rationale for the magnetic resonance imaging protocol for the knee. Osteoarthritis and cartilage. 2008;16(12):1433-1441
- Segal, Neil A., et al. "The Multicenter Osteoarthritis Study: opportunities for rehabilitation research." PM&R 5.8 (2013): 647-654.
- Kataoka, Hirokatsu, et al. "Would mega-scale datasets further enhance spatiotemporal 3D CNNs?." arXiv preprint arXiv:2004.04968 (2020).
- Kingma, Diederik P., and Jimmy Ba. "Adam: A method for stochastic optimization." arXiv preprint arXiv:1412.6980 (2014).
Figures
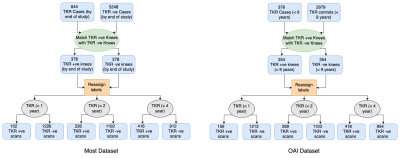
Figure 1: Cohort selection pipeline for the MOST and OAI datasets
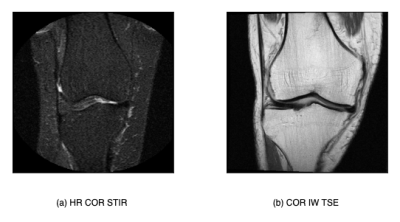
Figure 2: Comparison of a slice from the COR IW TSE contast (OAI dataset) and COR STIR contrast (MOST dataset)
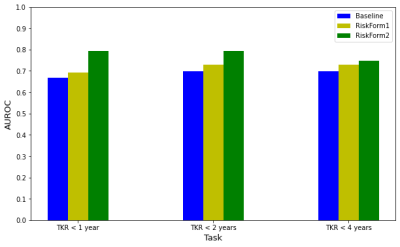
Figure 3: AUROC achieved by the models on TKR-positive knees only (< 9 years in OAI cohort). Note that the TKR < 1 year, < 2 years, and < 4 years labels for the images in this set can still be negative.
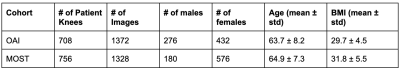
Table 1: Distribution of patients on the OAI and MOST sets
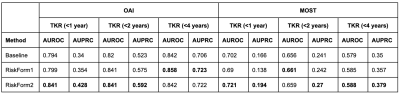
Table 2: Summary of Results. The AUROCs and AUPRCs computed on the OAI and MOST dataset on the three tasks TKR prediction <1 year, < 2 years and < 4 years
DOI: https://doi.org/10.58530/2023/1891