1881
Total Knee Replacement Prediction using Twin Class Distribution Estimation
Chaojie Zhang1, Shengjia Chen1, Haoxu Huang2, Haresh Rengaraj Rajamohan3, Jungkyu Park1, Noah Kasmanoff1, Kyunghyun Cho3, Gregory Chang1, Richard Kijowski1, and Cem M. Deniz1
1Department of Radiology, New York University Langone Health, New York, NY, United States, 2Courant Institute of Mathematical Sciences, New York University, New York, NY, United States, 3Center for Data Science, New York University, New York, NY, United States
1Department of Radiology, New York University Langone Health, New York, NY, United States, 2Courant Institute of Mathematical Sciences, New York University, New York, NY, United States, 3Center for Data Science, New York University, New York, NY, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence
Our study implemented the self-supervised learning method, Twin Class Distribution Estimation, with unlabeled knee MR images. The self-supervised pretraining improves the downstream analysis in predicting total knee replacement within 9 years using labeled knee MR images. The self-supervised features are shown to be efficient classifiers in TKR prediction.Introduction
Supervised learning with convolutional neural networks (CNNs) has shown great success in the classification of labeled medical images1-7. However, traditional supervised learning methods usually work with a large number of labeled data. The insufficiency of qualified labeled data degrades the model performance8-10. Recently, self-supervised learning was proposed to enable artificial intelligence systems to learn from unlabeled sample data. A mass of unlabeled images can be used to pretrain the deep learning model, and the model is subsequently fine-tuned using a small number of labeled image data11-12.Osteoarthritis (OA) is a degenerative joint disease and the most common reason leading to total knee joint replacement (TKR). It’s important and challenging to identify whether patients will progress to undergo the TKR before potential disease-modifying therapies can be effectively developed. Twin Class Distribution Estimation (TWIST) is a self-supervised learning method with siamese network architecture and has been proven to achieve better performance on ImageNet13. In our work, we applied TWIST in TKR prediction using knee MR images from the Osteoarthritis Initiative (OAI) dataset14.
Methods
DatasetSagittal intermediate-weighted turbo spin echo with fat suppression (SAG IW TSE FS) images (TE = 30 ms, TR = 3200 ms, FOV = 160 mm, Slice Thickness = 3.0 mm, Inplane Resolution = 0.357 mm × 0.511 mm, Bandwidth = 248 Hz/pixel, Matrix Size = 448 × 448 × 37) from the Osteoarthritis Initiative dataset14 were used in the study. Input of our deep learning models are 384 × 384 x 36 matrices randomly cropped from the MR images. All TWIST models are pretrained on the training set of 2800 unlabeled samples and the validation set of 286 unlabeled samples in the unsupervised learning task. In the transfer learning, we fine-tuned the TWIST model on a case-control cohort of 612 labeled training samples and 94 testing samples. The samples were identified as individuals who underwent a TKR within 9 years from the baseline and controls who didn't undergo a TKR within 9 years15. The matching was performed based on age, body mass index, ethnicity and sex.
Models and Training
The TWIST model generates twin class distributions of two augmented images using a softmax layer. By scattering samples into different pseudo-classes, the modified TWIST could distinguish different samples and learn representation from two unlabeled augmented MR images.
In pretraining, a couple of augmented images are generated from a 3D MR image and then fed into a siamese neural network composed of a 3D CNN, TSE-model backbone16. The outputs of the neural network are two probability distributions over pseudo categories. Figure 1 shows the network architecture and loss function of the modified TWIST. We trained separate models for the class of 8,32,128,512,1024 for 500 epochs with batch size of 16. Some routine data augmentation techniques are applied to the input MR images, including random flip, rescaling, rotation, Gaussian blur, and crop. All TWIST models are trained using SGD optimizer with layer-wise adaptive rate scaling.
To evaluate the self-supervised pretraining methods, we use the pretraining features as k-nearest neighbors (k-NN) classifiers in TKR prediction. The models are compared with other state-of-the-art self-supervised models, Barlow Twins17, BYOL18, and SimSiam19, trained using SGD optimizer with linear learning rate warmup of 50 epochs, and cosine learning rate decay. After pretraining, we fine-tuned the TWIST model with 612 labeled samples using six-fold cross-validation to predict whether individuals will undergo a TKR within 9 years or not. The models are compared with the purely supervised training model that was initialized randomly. We apply Gradient-weighted Class Activation Mapping (Grad-CAM)20 to highlight the important region in the knee MR images for the fine tuning model’s classification decision.
Results
Figure 2 shows the classification accuracy (in percentage) on downstream supervised learning tasks across hyperparameter k in the k-NN algorithm. The TWIST model trained by different target classes in the pretraining outperforms other self-supervised learning methods (Barlow Twins, BYOL, SimSiam) by 7.84%, 11.76%, 8.82%, and the supervised model trained with 612 labeled samples by 6.04%.Table 1 shows the transfer learning results for TKR prediction. The results indicate that TWIST outperforms supervised and other self-supervised learning methods, achieving an average AUC of 0.86 on six-fold cross-validation and 0.84 on the test set.
Figure 3 shows pseudo-classes predictions from TWIST. When TWIST learns representation by classifying, the output predictions are deterministic and the classes are diverse from each other. The predictions from two augmented images are consistent, indicating that TWIST learns representation by classifying two views into the same class.
For the case that will undergo a TKR within 9 years, we find the Grad-CAM highlights the joint region of the knee MR images (Figure 4).
Conclusion
We applied a novel self-supervised method TWIST with knee MR images to predict TKR. TWIST is able to classify a couple of augmented images into different categories without labels in pretext. It achieves better performance in k-NN classification and fine-tuning for TKR prediction, compared with supervised and other self-supervised learning methods. The visual interpretation of the fine-tuned model shows the potential of applying TWIST in clinical decision support.Acknowledgements
This work was supported in part by NIH grant R01 AR074453, and was performed under the rubric of the Center for Advanced Imaging Innovation and Research (CAI2R, www.cai2r.net), a NIBIB Biomedical Technology Resource Center (NIH P41 EB017183).References
- Esteva, Andre, et al. "Dermatologist-level classification of skin cancer with deep neural networks." nature 542.7639 (2017): 115-118.
- Sirinukunwattana, Korsuk, et al. "Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images." IEEE transactions on medical imaging 35.5 (2016): 1196-1206.
- Shin, Hoo-Chang, et al. "Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning." IEEE transactions on medical imaging 35.5 (2016): 1285-1298.
- Rajkomar, Alvin, et al. "High-throughput classification of radiographs using deep convolutional neural networks." Journal of digital imaging 30.1 (2017): 95-101.
- Shen, Wei, et al. "Multi-scale convolutional neural networks for lung nodule classification." International conference on information processing in medical imaging. Springer, Cham, 2015.
- Korolev, Sergey, et al. "Residual and plain convolutional neural networks for 3D brain MRI classification." 2017 IEEE 14th international symposium on biomedical imaging (ISBI 2017). IEEE, 2017.
- Roth, Holger R., et al. "Anatomy-specific classification of medical images using deep convolutional nets." 2015 IEEE 12th international symposium on biomedical imaging (ISBI). IEEE, 2015.
- Zhang, Jianpeng, et al. "Medical image classification using synergic deep learning." Medical image analysis 54 (2019): 10-19.
- Litjens, Geert, et al. "A survey on deep learning in medical image analysis." Medical image
- Cai, Lei, Jingyang Gao, and Di Zhao. "A review of the application of deep learning in medical image classification and segmentation." Annals of translational medicine 8.11 (2020).
- Xie, Zhenda, et al. "Simmim: A simple framework for masked image modeling." Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2022.
- Azizi, Shekoofeh, et al. "Big self-supervised models advance medical image classification." Proceedings of the IEEE/CVF International Conference on Computer Vision. 2021.
- Wang, Feng, et al. "Self-supervised learning by estimating twin class distributions." arXiv preprint arXiv:2110.07402 (2021).
- Peterfy, C.G., M.D., Ph.D, Schneider E, Ph.D, Nevitt M, Ph.D. The osteoarthritis initiative: Report on the design rationale for the magnetic resonance imaging protocol for the knee. Osteoarthritis and cartilage. 2008;16(12):1433-1441.
- Leung, Kevin, et al. "Prediction of total knee replacement and diagnosis of osteoarthritis by using deep learning on knee radiographs: data from the osteoarthritis initiative." Radiology 296.3 (2020): 584.
- Wang, Tianyu, et al. "Total knee replacement prediction using structural MRIs and 3D convolutional neural networks." International Conference on Medical Imaging with Deep Learning--Extended Abstract Track. 2019.
- Zbontar, Jure, et al. "Barlow twins: Self-supervised learning via redundancy reduction." International Conference on Machine Learning. PMLR, 2021.
- Chen X, He K. Exploring simple siamese representation learning[C]//Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2021: 15750-15758.
- Grill J B, Strub F, Altché F, et al. Bootstrap your own latent-a new approach to self-supervised learning[J]. Advances in neural information processing systems, 2020, 33: 21271-21284.
- Selvaraju, Ramprasaath R., et al. "Grad-cam: Visual explanations from deep networks via gradient-based localization." Proceedings of the IEEE international conference on computer vision. 2017.
Figures
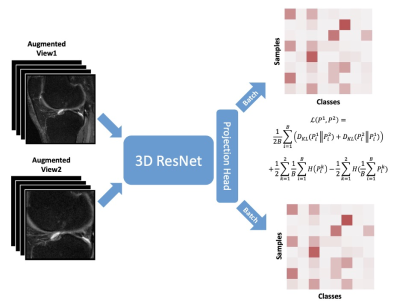
Figure 1: Network architecture of TWIST for 3D knee MR images.
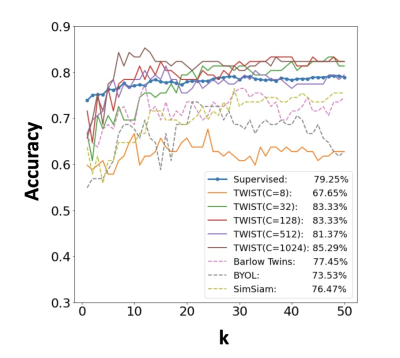
Figure 2: Total knee replacement prediction accuracy of k-nearest neighbors algorithm using supervised training and self-supervised pretraining features.
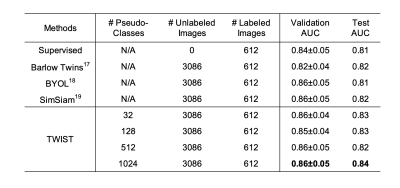
Table 1: Fully-supervised fine-tuning results. We compare different classes.

Figure 3: Probability distribution for two views from different augmentations. The number of samples (rows) is set to be the same as the class number (columns).
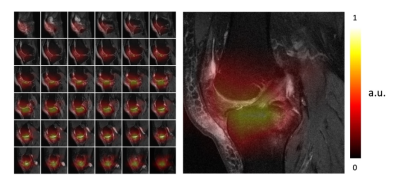
Figure 4: Gradient-weighted Class Activation Mapping for model interpretation.
DOI: https://doi.org/10.58530/2023/1881