1878
Deep Learning Assisted Diagnosis of Prostate Cancer: Using a Multi-scale Neural Network Based on Points of Interest1Department of Magnetic Resonance, Lanzhou University Second Hospital, Lanzhou, China, 2Department of Magnetic Resonance, Xijing Hospital, Xi'an, China, 3Department of Radiology, Xijing Hospital, Xi'an, China, 4Philips Healthcare, Xi’an, China
Synopsis
Keywords: Prostate, Machine Learning/Artificial Intelligence, Spatial Transformer Network, Transfer learning
In this study, we propose a method to predict clinically significant state cancer based on MRI points of interest (POI) and classification network with multi-mode and multi-scale. Instead of the traditional method of manual delineation region-of-interest (ROI) to assist prediction, our method utilizes multi-scale input combined with Spatial Transformer Network (STN) to automatically adjust the adjust the scale of interest. This work also explored the possibility of predicting the grade of prostate cancer in a small amount of data using the method of transfer learning. Experiments show that this method has high prediction performance.Introduction
Prostate cancer is the most common cancer diagnosis made in men and the fifth leading cause of death worldwide. Early diagnosis is very important for future treatment. Current gold standard screening and grading method for prostate cancer is transrectal ultrasound (TRUS)-guided biopsy and the Gleason grading system. However, the (TRUS)-guided biopsy is an invasive technique that acquires tissue specimens from prostate gland. Computer-aided diagnostic allow us to differentiate different prostate diseases non-invasively. Previous studies used machine learning method to predicted whether the patient was prostate cancer, significant prostate cancer (Gleason score ≥ 2), clinically significant prostate cancer(Gleason score ≥ 7) or not respectively[1-3]. Vente et al.,[4] made further experiments on the multi classification problem and predicted Gleason Grade Group (GGG). Previous researchers supported by a large amount of data, proved that the ROI aided prediction method on MRI is successful in prostate cancer. However, a large amount of data needs to be collected for a long time, and it is very laborious to manually mark ROI. Therefore, this paper uses pre training and transfer learning to explore the classification of prostate cancer lesion level under small data sets and few artificial assistances (marking points of interest).Methods
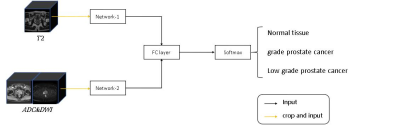
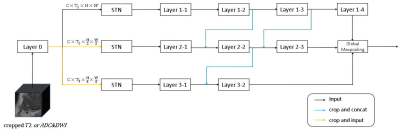
MR data and pathological information of 117 patients with prostate diseases were obtained from the Xijing Hospital and PROSTATEx-2. The 18 patients collected at the First Affiliated Hospital of Airforce Medical University underwent MR scan by a 3.0T MR (Ingenia CX, Philips Healthcare, the Netherlands) with a 16-channel torso coil. T2 weighted images and diffusion weighted images were include in the MR examination. Apparent diffusion coefficient (ADC) maps were automatically calculated by the MR manufacturer. Then, we propose a 3D convolutional neural network based on points of interest to differentiate the grade of prostate cancer. Specifically, we take the local 3D MRI image patch centered on the point of interest as the input. As shown in Figure 1, there are two parts in our network, one part of the input is T2 weighted high-resolution images, and the other part is DWI low-resolution images and the corresponding ADC maps. The two parts have similar network architectures. In the network design (Figure 2), to better adapt to the size of different prostate cancer lesions, the input MRI images are first simply convolved, and then the feature map will be divided into three images of different scales and input into three different networks. In the input layer of the three sub-networks, we also add the Spatial Transformer Network (STN), which enables the network to adaptively adjust the region of interest of the image.We first pre trained this network under the PROSTATEx-2 dataset, which includes 99 patients and 112 lesions. We split the training set, verification set and test set according to the patients with a ratio of 7:1.5:1.5. Before entering the network, the image will be enhanced by randomly cropping, stretching and gamma adjustment. After the pre training, the network will be transferred to the local data for fine tuning. The local data includes 18 patients and 22 lesions, and we marked a non-lesion point of interest on each patient. Set the ratio of training set and test set to 7:3 in local data. The training process uses the same data enhancement operations as pre training.
Results
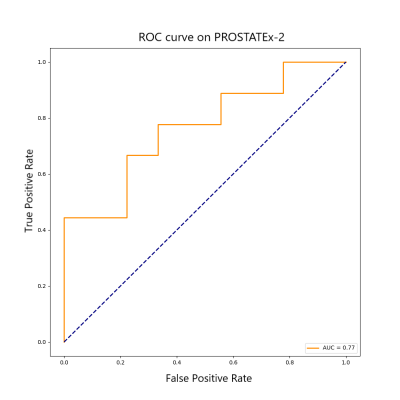
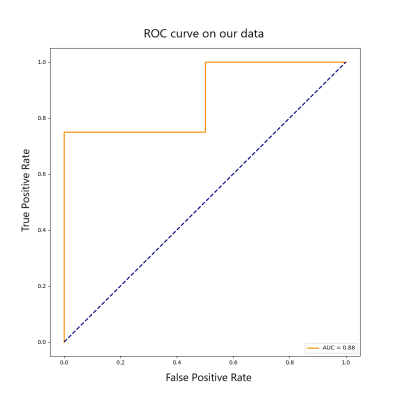
In the pre trained PROSTATEx-2 data, the model is verified by the validation set using early stopping, and finally the AUC of the test set is 0.77. In the fine-tuning training of local data, the AUC in predicting whether a patient has clinically significant PCa is 0.88, and the Kappa coefficient in the three classifications containing normal cases is 0.7. The specific ROC curve is shown in Figures 3 and 4.Discussion
Traditional practices require a large amount of data and long -term local training, and the ROI marked by different clinicians is inconsistent, which possess subjectivity. The research in this paper has improved the experimental method based on the above problems. The difficulty of this study is not to use the region of interest, which makes the network unable to determine the scale of the lesion. Therefore, we used multi-scale images and STN methods to enable the neural network to automatically adjust the region of interest. Of course, multi branch networks will increase the number of parameters. The basic block used in this paper is shown in Figure 5, we replaced the 3x3x3 convolution with 3x1x1 convolution and 1x3x3 convolution. This operation can be seen as Deepwise convolution with slice scale information, and can reduce the number of parameters of our model by about 50%.Conclusion
This paper explores the prediction of prostate cancer in MRI with small amount of data. The experiment proves that using pre training network and a small number of data sets to fine tune can achieve a higher prediction effect. Moreover, our model achieves a high prediction effect with simple manual assistance. The proposed methods and models are promising tool to differentiate different grade of prostate cancer and may be extended to other disease prediction problems.Acknowledgements
NoneReferences
[1]. Song, Yang et al. “Computer-aided diagnosis of prostate cancer using a deep convolutional neural network from multiparametric MRI.” Journal of magnetic resonance imaging : JMRI vol. 48,6 (2018): 1570-1577. doi:10.1002/jmri.26047
[2]. Schelb, Patrick et al. “Classification of Cancer at Prostate MRI: Deep Learning versus Clinical PI-RADS Assessment.” Radiology vol. 293,3 (2019): 607-617. doi:10.1148/radiol.2019190938
[3]. Mehta, Pritesh et al. “Computer-aided diagnosis of prostate cancer using multiparametric MRI and clinical features: A patient-level classification framework.” Medical image analysis vol. 73 (2021): 102153. doi:10.1016/j.media.2021.102153
[4]. Vente, Coen de et al. “Deep Learning Regression for Prostate Cancer Detection and Grading in Bi-Parametric MRI.” IEEE transactions on bio-medical engineering vol. 68,2 (2021): 374-383. doi:10.1109/TBME.2020.2993528
[5]. K. He, X. Zhang, S. Ren and J. Sun, "Deep Residual Learning for Image Recognition," 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2016, pp. 770-778, doi: 10.1109/CVPR.2016.90
[6]. Jaderberg, Max, Karen Simonyan, and Andrew Zisserman. "Spatial transformer networks." Advances in neural information processing systems 28 (2015)
[7]. Trebeschi S ,Drago S G ,Birkbak N J ,et al.Predicting Response to Cancer Immunotherapy using Non-invasive Radiomic Biomarkers. Annals of Oncology, 2019.
[8]. Geert Litjens, Oscar Debats, Jelle Barentsz, Nico Karssemeijer, and Henkjan Huisman. "ProstateX Challenge data", The Cancer Imaging Archive (2017). DOI: 10.7937/K9TCIA.2017.MURS5CL
[9]. Litjens G, Debats O, Barentsz J, Karssemeijer N, Huisman H. "Computer-aided detection of prostate cancer in MRI", IEEE Transactions on Medical Imaging 2014;33:1083-1092. DOI: 10.1109/TMI.2014.2303821
[10]. Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository, Journal of Digital Imaging, Volume 26, Number 6, December, 2013, pp 1045-1057. DOI: 10.1007/s10278-013-9622-7