1827
Self-navigated free-breathing ZTE lung imaging1GE HealthCare, Little Chalfont, Amersham, United Kingdom, 2ASL Europe, GE HealthCare, Munich, Germany, 3Department of Radiology, University of Cambridge, Cambridge, United Kingdom, 4University of Cambridge School of Clinical Medicine, Royal Papworth Hospital, Cambridge, United Kingdom, 5Medical Radiation Physics, Lund University, Lund, Sweden, 6Neuroimaging, Institute of Psychiatry, Psychology, and Neuroscience, King's College London, London, United Kingdom
Synopsis
Keywords: Motion Correction, Motion Correction, Self-navigation
Here we propose a self-navigated technique, based on the extraction of respiratory motion estimates using interleaved spiral phyllotaxis trajectories in 3D radial zero echo time (ZTE) acquisitions. These are particularly well-suited to capture the short T2* signal, characteristic of lung parenchyma, and have high acquisition efficiency allowing to generate fast temporal resolution navigators. The self-navigation technique worked robustly for different respiratory patterns and on 1.5T and 3.0T fields strengths, obtaining high quality images comparable to the ones obtained with bellows-gating.Introduction
Clinical lung imaging workflows typically use short breath-hold acquisitions or respiratory triggering to reduce motion artifacts. However, free-breathing acquisitions are desirable for increased patient comfort and allow for high-resolution volumetric images in a fixed scan time.Breathing motion is usually corrected using navigators or auxiliary motion sensors such as respiratory bellows or external trackers. The former decrease scan efficiency and interfere with the imaging process, while the latter complicate workflow, increase cost, require cross-calibration and may pick-up additional motions. In particular, widely available respiratory bellows cannot provide absolute displacements, their signal tends to drift and are often poorly coupled with the diaphragmatic motion, which may result in inaccurate respiratory motion estimation of the volume of interest.
To address these issues, here we propose a self-navigated technique, based on the extraction of respiratory motion estimates using interleaved spiral phyllotaxis trajectories in 3D radial ZTE acquisitions. These are particularly well-suited to capture the short T2* signal, characteristic of lung parenchyma, and have high acquisition efficiency allowing to generate fast temporal resolution navigators.
Methods
4 volunteers were recruited with an institutionally-approved IRB protocol. Free-breathing scans of the lung region were performed on GE 1.5T Signa Artist and a GE 3.0T Signa Premier (GE Healthcare, Chicago, IL) using a 3D radial ZTE sequence [2] (FOV=38.4 mm3, res=1.25-1.5mm3, FA=1deg, BW=±83.3kHz, NEX=8, time=8-10 minutes). Respiratory bellows were used to capture the respiratory pressure waveform (RWBELLOWS).Low resolution images (3.2mm3) were generated using spiral phyllotaxis interleaves. Each frame was reconstructed from 1024 spokes distributed over 4 segments, with a time resolution Δt=1.18s. Principal Component Analysis (PCA) of the dominant motion component was used to generate the data-driven self-navigated respiratory waveform (RWPCA).
Three sets of 3D ZTE images were reconstructed using 3D gridding: non-gated images (1), gated images using the respiratory waveforms RWBELLOWS (2) and RWPCA (3). End-inspiration and end-expiration phases were selected by thresholding the amplitude histogram. Additionally, we acquired 3 datasets with a prototype that retrospectively selects the quiescent phase by maximizing the gating weights in the quiescent phase bin [3].
The spiral phyllotaxis trajectory golden-angle distribution allows to retrospectively shorten the datasets while keeping approximately uniform sampling density. 4 subsampled datasets (75%,50%,25%,12.5%) were generated from the fully-sampled data to determine the shortest acquisition that preserves fine pulmonary vasculature, after measuring the dice similarity coefficient (DSC) of ROIs covering the thinnest branches in these images.
Results
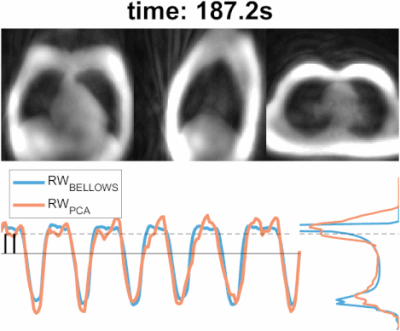
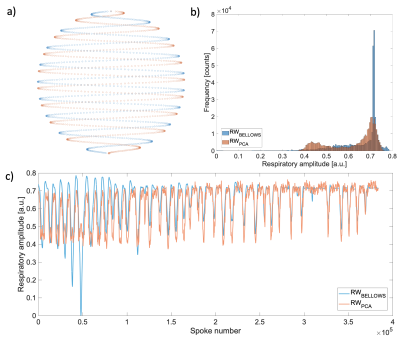
The animated Figure 1 shows low resolution images reconstructed from individual interleaves of the spiral phyllotaxis trajectory, schematically shown in Figure 2a. Figure 1 (bottom left) and Figure 2b show the self-gated derived respiratory waveform of two volunteers and the corresponding signal acquired with the respiratory bellows. Note that the respiratory bellows waveforms tend to show larger drift than the PCA derived waveforms.Figure 1 (bottom right) and Figure 2c present the amplitude histogram of the waveforms; in the most challenging cases the separation between peaks is not broad, but thresholding allows to gate two distinct respiratory states and reject the least coherent data from each bin. Furthermore, Figure 2b shows how the self-gated waveform recovers an end-expiration peak that is spread across the belt waveform. Overall, the PCA method generates respiratory waveforms that are well correlated (correlation coefficient >= 0.8) with the bellows signal.
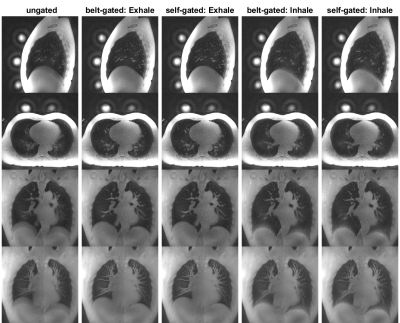
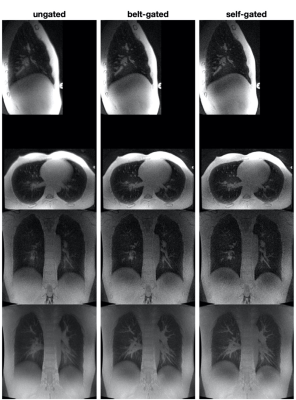
Figure 3 illustrates ungated images, bellows-gated and self-gated images at end-expiration and end-inspiration. As shown in the histograms in Figures 1 and 2c, end-expiration is the respiratory state where most of the data is usually acquired, and thus provides the sharpest images. Maximum Intensity Projection (MIP) was applied to these images to showcase the fine structure of lung vasculature.
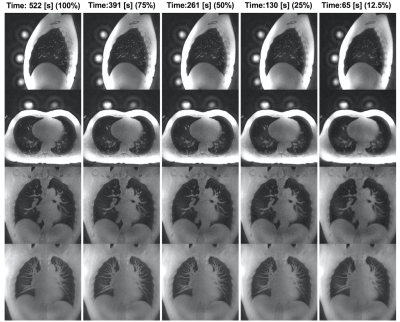
Figure 4 shows the same dataset as Figure 3, reconstructed with different fractions of the data. We calculated the DSC in 4 different regions covering the thinnest branches. The mean DCS values are 0.93±0.02, 0.87±0.04, 0.77±0.05 and 0.57±0.13 for the 75%, 50%, 25% and 12.5% subsample datasets, respectively.
Figure 5 presents a 1.5T dataset reconstructed on an empirically optimized quiescent phase, which depends on the respiratory signal, and provide good quality results for both belt-gated and self-gated waveforms.
Discussion
Here we show the feasibility of generating respiratory waveforms using interleaved spiral phyllotaxis trajectories in 3D radial ZTE acquisitions, without the need of external hardware or navigator lengthened acquisitions. The self-navigation technique worked robustly for different respiratory patterns, obtaining high quality images comparable to the ones generated with the bellows-gating.Longer acquisitions (acquisition time ~8.7mins) were chosen to demonstrate the feasibility of reconstructing retrospectively end-inspiration and end-expiration phases. We determined empirically that we can limit the acquisition to ~50% of the data (acquisition time ~4.35mins) while preserving the thinnest pulmonary vasculature. Shorter scans also tend to reduce the likelihood of drift and bulk motion, helping to improve data consistency.
The technique was tested at 1.5T and 3T field strengths using the same protocol to facilitate the comparison of the results. However, more efficient protocols will be explored, as well as the possibility to generate multiple-phase images using soft-gated techniques.
Additional image quality improvements can be achieved applying a customized denoising DL model to sharpen the images, which could be used to further reduce the acquisition time while maintaining high SNR.
Acknowledgements
This research was supported by the NIHR Cambridge Biomedical Research Centre (BRC-1215-20014). The views expressed are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care.References
[1] Piccini, D., Littmann, A., Nielles-Vallespin, S. and Zenge, M.O. (2011), Spiral phyllotaxis: The natural way to construct a 3D radial trajectory in MRI. Magn. Reson. Med., 66: 1049-1056. https://doi.org/10.1002/mrm.22898.
[2] Liu X, Gómez PA, Solana AB, Wiesinger F, Menzel MI, Menze BH. Silent 3D MR sequence for quantitative and multicontrast T1 and proton density imaging. Phys Med Biol. 2020 Sep 16;65(18):185010. doi: 10.1088/1361-6560/aba5e8. PMID: 32663809.
[3] Bae K, Jeon KN, Hwang MJ, Lee JS, Park SE, Kim HC, Menini A. Respiratory motion-resolved four-dimensional zero echo time (4D ZTE) lung MRI using retrospective soft gating: feasibility and image quality compared with 3D ZTE. Eur Radiol. 2020 Sep;30(9):5130-5138. doi: 10.1007/s00330-020-06890-x. Epub 2020 Apr 24. PMID: 32333146.
Figures