1824
Fast, model-based, and navigator-free retrospective motion correction for non-Cartesian fMRI
Guanhua Wang1, Shouchang Guo2, Jeffrey A. Fessler2, and Douglas C. Noll1
1Biomedical Engineering, University of Michigan, Ann Arbor, MI, United States, 2EECS, University of Michigan, Ann Arbor, MI, United States
1Biomedical Engineering, University of Michigan, Ann Arbor, MI, United States, 2EECS, University of Michigan, Ann Arbor, MI, United States
Synopsis
Keywords: Motion Correction, Motion Correction
This abstract presents a retrospective motion correction method for fMRI. The method alternates between motion estimation and motion-informed model-based reconstruction. Compared to registration-based correction, this approach resolves intra-frame, inter-shot motion without additional navigators. The open-source GPU-based implementation enables efficient correction/reconstruction for large-scale non-Cartesian fMRI data. With prospective experiments, we demonstrate that our approach outperformed retrospective registration by providing higher-resolution images with reduced false positives in activation maps.Introduction
Motion correction is important for fMRI due to the long acquisition time. Currently, registration-based retrospective correction is standard in post-processing pipelines [1]. Typically, this approach estimates the motion parameters for the whole volume at a certain time frame. For high-resolution multi-shot fMRI, the intra-frame motion between shots may impede these image-based approaches and cause blurring. We propose an iterative approach for fine-grained motion correction and reconstruction. By iterating between motion estimation and model-based reconstruction, this method leads to improved motion correction compared to single-pass registration. The implementation uses multi-GPU optimization to accelerate the computation.Methods
The proposed method alternates between motion estimation (via registration) and motion-informed model-based reconstruction to synergistically improve both. The first step reconstructs a ‘crude’ image for each acquisition shot. We used a ‘data-sharing’ (multi-shot) reconstruction as the initialization to avoid strong aliasing artifacts. The next step estimates rigid motion by registration (gradient correlation as the metric). Then the method updates the k-space data and corresponding system matrices of each shot (including the sampling trajectory and coil sensitivity and B0 maps) according to the motion parameters.The model-based reconstruction usually involves the following cost function $$$\hat{x}=\underset{x}{\arg \min }\|A x-y\|_{2}^{2}+\mathrm{R}(x)$$$, where $$$A$$$ is the system matrix. Here the system matrix is a stack of system matrices of different shots $$A = \begin{bmatrix} A_{\mathrm{shot1}} \\ A_{\mathrm{shot2}} \\ \vdots \\ A_{\mathrm{shotn}} \end{bmatrix},$$ to accommodate different trajectories and shifted sensitivity/B0 maps. R(x) denotes the regularization terms, for example, a Tikhonov regularizer.
Compared to standard reconstruction, the proposed reconstruction method requires more NUFFTs for multi-shot acquisition. The implementation uses the GPU-based MIRTorch [2] and torchkbnufft [3] for efficient computation.
Experiments
We tested the proposed motion correction with the oscillating steady state (OSS) fMRI [4], a novel high-SNR BOLD fMRI method. The sampling trajectory used the rotation EPI (REPI) [5], as shown in Fig. 1. The FOV is 20×20×3.2 cm3, with 2 mm3 isotropic resolution. The TR is 16 ms, and TE is 7.4 ms. We used 10 shots and 10 OSS ‘fast time’ acquisitions to reconstruct a single time frame. In the phantom experiment, we cyclically rotated a cylinder water phantom. In the in-vivo task-based study, a volunteer performed the finger-tapping task, and rotated the head to mimic motion. The baseline registration used the standard MCFLIRT function in the FSL toolbox [1]. The reconstruction algorithm was CG-SENSE reconstruction (roughness penalty, 20 iterations) and used Nvidia RTX8000 GPUs.Results and Discussion
Fig. 2 shows the results of the phantom experiment. Compared with MCFLIRT, our method effectively reduced the blurring caused by the intra-frame motion. Fig. 3 displays the in-vivo reconstruction results, where the proposed method reduced blurring. Fig. 4 shows activation maps calculated from MCFLIRT and the proposed method. MCFLIRT falsely showed activation in the skull due to motion, whereas the proposed method accurately reflected voxels in the motor cortex.Our approach with the multi-GPU optimization significantly improves computation efficiency: the reconstruction time of using 3 GPUs was <1hr, whereas an 18-core CPU calculation took more than 20 hr (for 60 time frames).
We showed an application to fMRI in this abstract. It may also benefit other applications, such as diffusion and MR fingerprinting.
Acknowledgements
This work is supported by NIH Grants R01 EB023618.References
[1] Jenkinson, M., Bannister, P., Brady, M., & Smith, S. (2002). Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage, 17(2), 825-841.
[2] https://github.com/guanhuaw/MIRTorch
[3] https://github.com/mmuckley/torchkbnufft
[4] Guo, S., & Noll, D. C. (2020). Oscillating steady‐state imaging (OSSI): A novel method for functional MRI. Magnetic resonance in medicine, 84(2), 698-712.
[5] Rettenmeier, C. A., Maziero, D., & Stenger, V. A. (2022). Three dimensional radial echo planar imaging for functional MRI. Magnetic Resonance in Medicine, 87(1), 193-206.
Figures

Figure 1. Workflow of the proposed method.
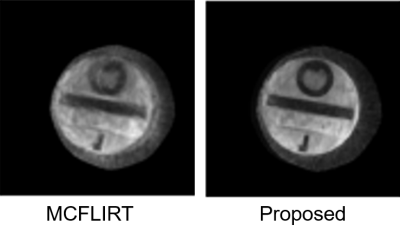
Figure 2. Results of the phantom experiment. In the presence of strong motion, the registration-based method (MCFLIRT) cannot resolve the blurring caused by the inter-shot movement.
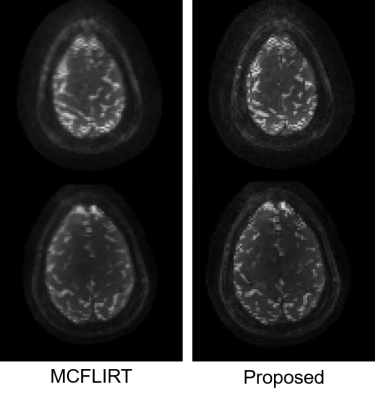
Figure 3. Results of the in-vivo experiment. Two slices of the same time point are displayed, corrected by different methods. Similar to the phantom experiment, the proposed method effectively reduced the blurring.
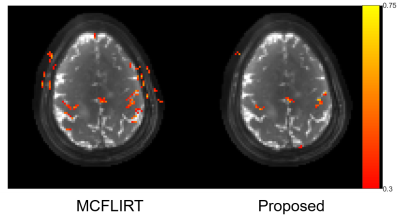
Figure 4. Activation maps for the two motion correction methods. MCFLIRT led to scattered false positives due to motion, while the proposed iterative method reflected accurate correlations of the motor cortex.
DOI: https://doi.org/10.58530/2023/1824