1748
Role of Diffusion Magnetic Resonance Imaging in Brain Meningioma1Chitwan Medical College and Teaching Hospital, Chitwan, Nepal, 2Sharda University, Greater Noida, India, 3Indiana University School of medicine, Stark Neuroscience, IndianaPolis, IN, United States, 4Indiana University School of medicine, IndianaPolis, IN, United States
Synopsis
Keywords: Tumors, Brain
Meningioma as an extra-axial tumor of meninges is difficult to interpret within brain interface using normal Magnetic Resonance Imaging sequence protocol. Recently, Various advancement in diffusion Magnetic Resonance Imaging (dMRI) techniques such as DTI (Diffusion Tensor Imaging), DKI (Diffusion Kurtosis Imaging) and NODDI (Neurite Orientation Dispersion and Density Imaging) has been utilized as an emerging diagnostic tool to measure the degree of mobility of water molecules within biologic tissue and investigate micro-structural integrity of the brain tissue. Thus, we aim to differentiate meningiomas from normal brain parenchyma using DTI, DKI and NODDI metrics.
Introduction
Magnetic Resonance Imaging (MRI) is a non-invasive diagnostic imaging modality that is desirable to predict aggressiveness of meningiomas because it would help foresee tumor recurrence and improve postoperative tumor management 1. Meningiomas are the extra axial tumors and represent the most common tumor of meninges that is easy to diagnose with characteristic imaging findings in conventional MRI. However, the ability of conventional MRI to differentiate meningioma from normal brain tissue remains uncertain. Some studies have reported that conventional MRI findings are not specific and reliable in identifying and distinguishing meningioma from normal brain parenchyma 2. Diffusion Magnetic Resonance Imaging (dMRI) is a powerful technique to investigate micro-structural integrity of the brain tissue 3. Lately, there has been various advancement in dMRI techniques such as DTI (Diffusion Tensor Imaging), DKI (Diffusion Kurtosis Imaging) and NODDI (Neurite Orientation Dispersion and Density Imaging) to differentiate and for early detection of meningioma from normal brain tissue. Therefore, the purpose of this study was to examine whether dMRI models known as DTI, DKI and NODDI could provide more accurate diagnosis in differentiating meningioma from normal brain.Methods
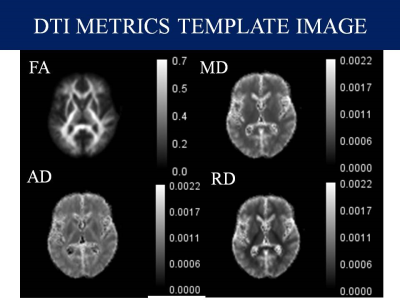
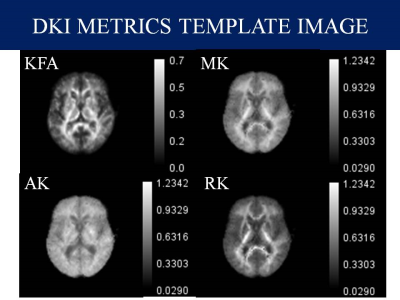
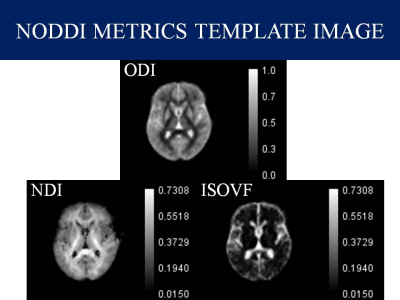
Data Collection: In this study, we included preoperative data of 8 Meningioma and 8 Normal controls available from the The Virtual Brain, an open-source neuroinformatic platform under the name Brain Tumor Connectomics 4. we included only the low grade meningioma whereas high grade meningiomas were excluded because of unclear tumor-brain interface.Data Analysis: Preprocessing of Diffusion MRI data was done using the combination of FSL and MRtrix3 in Bash (Shell scripting) in Ubuntu 20.04. First, raw diffusion MRI data were corrected for artifacts. In particular, the preprocessing of data include Denoising 5, Gibbs artifact removal 6, Top up 7 followed by the Eddy current correction 8, Bias field correction 9 and Brain mask estimation 10. DTI metrics; Fractional Anisotropy (FA), Mean Diffusivity (MD), Axial Diffusivity (AD) and Radial Diffusivity (RD) were extracted from MRtrix3 package 11, DKI metrics; Kurtosis Fractional Anisotropy (KFA), Mean Kurtosis (MK), Axial Kurtosis (AK) and Radial Kurtosis (RK) were calculated from DIPY Python package 12 and NODDI metrics Orientation Distribution Index (ODI), Neurite Dispersion Index (NDI) and Isometric Volume Fraction (ISOVF) from AMICO package 13. Region of Interest (ROIs) were drawn manually using ITK-Snap 14 and cystic components were avoided while estimating the solid region of the tumor. As shown in Figure 1-3, Various maps with calibration bars were created for all the DTI, DKI and NODDI metrics using ImageJ Application 15.
Statistical Analysis: SPSS, version 23.0 was used to perform statistical analysis for meningioma and normal participants. First, The Shapiro-Wilks test of normality was used to analyze the normal distribution assumption of the quantitative outcomes. Secondly, we compared the mean between meningioma patients and normal patients. Based on this outcome, an independent sample t Test was used in order to determine whether there is statistical evidence that the meningioma and normal patients’ means are significantly different and p<0.05 was considered as statistically significant. All the data were recorded as Mean±SD.
Results
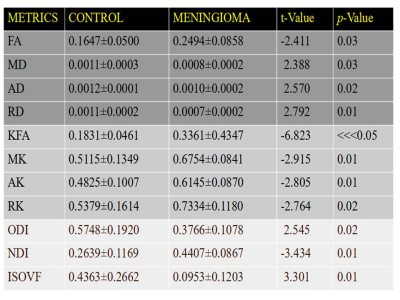
Shown in Table.1, Mean±SD and corresponding statistical analysis of DTI metrics show statistical significant difference between control and meningioma patients (p<0.05) among which FA metrics tends to increase in meningioma whereas MD, AD and RD tends to decrease in meningioma in compare to control group. Similarly, Statistical Analysis of DKI metrics show statistical significant difference between control and meningioma patients (p<0.05), all the DKI metrics show higher mean value in meningioma. Furthermore, Statistical Analysis of NODDI metrics show significant difference between control and meningioma patients (p<0.05).Discussion
In this study, our results demonstrated that dMRI metrics; DTI, DKI and NODDI enabled meningioma to differentiate from normal brain showing statistical significant difference (p<0.05). Thus, dMRI techniques are useful to differentiate meningioma from normal brain which is a limitation of conventional MRI technique 16. FA gives information about the directionality of water diffusion and shows tissue integrity 17. Our findings showed that FA was higher in meningioma as compared to normal patients (p=0.03), reflects the higher density and higher micro structural organization of the tumor. On the contrary, the results of our study showed that all DKI metrics were significantly higher in meningioma than control patients showing statistical significance (p<0.05). The increase of Kurtosis in meningioma probably reflects a higher degree of micro structural complexity within the tumor 18. DKI, which estimates the non-Gaussian diffusion distribution might characterize the heterogeneity of the micro environment and also has provided further insights into tissue characteristics 19. Similarly, Statistical Analysis of NODDI metrics show significant difference between control and meningioma patients (p<0.05), where ODI value tends to decrease in case of meningioma patients, whereas, NDI and ISOVF tends to increase in meningioma suggesting micro-structural tissue derangement's 20.Conclusion
Our study revealed that the advanced dMRI techniques, DTI, DKI and NODDI, were useful in differentiating meningioma from normal brain tissue. The distinctive ability of dMRI models has confirmed to be more accurate and sensitive for assessing and predicting meningioma. Thus, dMRI parameters can be used as a useful diagnostic tool when distinguishing meningioma from normal brain tissue.Acknowledgements
NoneReferences
1. Tang Y, Dundamadappa SK, Thangasamy S, Flood T, Moser R, Smith T, Cauley K, Takhtani D. Correlation of apparent diffusion coefficient with Ki-67 proliferation index in grading meningioma. AJR Am J Roentgenol. 2014 Jun 1;202(6):1303-8.
2. Karthigeyan M, Dhandapani S, Salunke P, Singh P, Radotra BD, Gupta SK. The predictive value of conventional magnetic resonance imaging sequences on operative findings and histopathology of intracranial meningiomas: a prospective study. Neurology India. 2019 Nov 1;67(6):1439.
3. Xiao X, Kong L, Pan C, Zhang P, Chen X, Sun T, Wang M, Qiao H, Wu Z, Zhang J, Zhang L. The role of diffusion tensor imaging and tractography in the surgical management of brainstem gliomas. Neurosurgical Focus. 2021 Jan 1;50(1):E10.
4. Aerts, H and Marinazzo, D (2018). BTC_preop. OpenNeuro. [Dataset] doi: doi:10.18112/openneuro.ds001226.v5.0.0
5. Veraart J, Novikov DS, Christiaens D, Ades-Aron B, Sijbers J, Fieremans E. Denoising of diffusion MRI using random matrix theory. Neuroimage. 2016 Nov 15;142:394-406.
6. Kellner E, Dhital B, Kiselev VG, Reisert M. Gibbs‐ringing artifact removal based on local subvoxel‐shifts. Magnetic resonance in medicine. 2016 Nov;76(5):1574-81.
7. Andersson JL, Skare S, Ashburner J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. Neuroimage. 2003 Oct 1;20(2):870-88.
8. Andersson JL, Sotiropoulos SN. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage. 2016 Jan 15;125:1063-78.
9. Tournier JD, Smith R, Raffelt D, Tabbara R, Dhollander T, Pietsch M, Christiaens D, Jeurissen B, Yeh CH, Connelly A. MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. Neuroimage. 2019 Nov 15;202:116137.
10. Smith SM. Fast robust automated brain extraction. Human brain mapping. 2002 Nov;17(3):143-55.
11. Westin, C.-F., Peled, S., Gudbjartsson, H., Kikinis, R. and Jolesz, F. A.. "Geometrical Diffusion Measures for MRI from Tensor Basis Analysis." Paper presented at the meeting of the ISMRM '97, Vancouver Canada, 1997.
12. Contributors D, Garyfallidis E, Brett M, Amirbekian BB, Rokem A, van der Walt S, Descoteaux M, Nimmo-Smith I. Dipy, a library for the analysis of diffusion MRI data. Frontiers in Neuroinformatics. 2014 Feb 21;8:8.
13. Daducci A, Canales-Rodríguez EJ, Zhang H, Dyrby TB, Alexander DC, Thiran JP. Accelerated microstructure imaging via convex optimization (AMICO) from diffusion MRI data. Neuroimage. 2015 Jan 15;105:32-44.
14. Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006 Jul 1;31(3):1116-28.
15. Rueden CT, Eliceiri KW. ImageJ for the next generation of scientific image data. Microscopy and microanalysis. 2019 Aug;25(S2):142-3.
16. Watts J, Box G, Galvin A, Brotchie P, Trost N, Sutherland T. Magnetic resonance imaging of meningiomas: a pictorial review. Insights into imaging. 2014 Feb;5(1):113-22.
17. Nagar VA, Ye JR, Ng WH, Chan YH, Hui F, Lee CK, Lim CC. Diffusion-weighted MR imaging: diagnosing atypical or malignant meningiomas and detecting tumor dedifferentiation. American Journal of Neuroradiology. 2008 Jun 1;29(6):1147-52.
18. Lin L, Bhawana R, Xue Y, Duan Q, Jiang R, Chen H, Chen X, Sun B, Lin H. Comparative analysis of diffusional kurtosis imaging, diffusion tensor imaging, and diffusion-weighted imaging in grading and assessing cellular proliferation of meningiomas. American Journal of Neuroradiology. 2018 Jun 1;39(6):1032-8.
19. Tietze A, Hansen MB, Østergaard L, Jespersen SN, Sangill R, Lund TE, Geneser M, Hjelm M, Hansen B. Mean diffusional kurtosis in patients with glioma: initial results with a fast imaging method in a clinical setting. American journal of neuroradiology. 2015 Aug 1;36(8):1472-8.
20. Pieri V, Sanvito F, Riva M, Petrini A, Rancoita PM, Cirillo S, Iadanza A, Bello L, Castellano A, Falini A. Along‐tract statistics of neurite orientation dispersion and density imaging diffusion metrics to enhance MR tractography quantitative analysis in healthy controls and in patients with brain tumors. Human brain mapping. 2021 Apr 1;42(5):1268-8
Figures