1690
Deep Learning-Based Automatic Segmentation of Non-contrast Coronary Magnetic Resonance Angiography Images1Department of Radiology, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China, 2Shukun (Beijing) Technology Co., Ltd, Beijing, China
Synopsis
Keywords: Heart, Cardiovascular
Coronary Magnetic Resonance Angiography (CMRA) is the only non-invasive coronary artery imaging method without radiation exposure and contrast media, and its application in clinical practice has been increasing. However, image post-processing for clinical diagnosis is time-consuming and requires expertise for radiologists. We proposed a three-dimensional U-Net-based automatic method for CMRA images by transfer learning from a pre-trained model of coronary computed tomography angiography(CCTA) to obtain accurate segmentation of coronary arteries.Introduction
Cardiovascular disease (CVD) is a leading cause of mortality and morbidity worldwide1. Patients with CVD are at risk of myocardial ischemia and consequent cardiac events2. Coronary magnetic resonance angiography(CMRA) is a non-invasive imaging method for assessing coronary stenosis. It has advantages over coronary computed tomography angiography(CCTA) because it does not require radiation exposure or the use of contrast media. However, due to the significant technical limitations of CMRA, including reduced spatial resolution, long acquisition time, and low signal-to-noise ratio, CMRA is not widely applied as CCTA3. These days, the use of deep learning techniques to improve image quality has provided new possibilities for the clinical application of CMRA4. However, image post-processing and vessel reconstruction need to be done by experienced radiologists, which is cumbersome and labor-intensive5. Therefore, we proposed a deep learning-based post-processing model of CMRA to automatically achieve coronary artery segmentation and reconstruction, facilitating the process of clinical diagnosis.Methods
Our study retrospectively included 1.5T non-contrast whole-heart CMRA images from 104 consecutive patients in 2 centers, divided into training and validation datasets. Scans with non-diagnostic image quality were excluded by manual inspection. In addition, we excluded patients with incomplete records and patients who had previously undergone percutaneous coronary intervention or coronary artery bypass grafting, or other cardiac procedures. 85 and 19 patients were randomly selected as the training and validation dataset, respectively. In this work, we applied transfer learning by transferring the vascular segmentation model of CCTA to CMRA, using a medium amount of training data to obtain a model with better performance. In our previous studies6,7, we proposed a DL-based automated algorithm for CCTA reconstruction, utilizing more than 500 cases from a multi-center dataset. Before training, aortas, coronary arteries, and plaques were labeled on each image by trained graders and checked by experts with more than five years of experience. Our proposed deep convolutional neural network adopts the improved three-dimensional U-Net architecture to establish the Growing Iterative Prediction Network (GIPN) model. The network can obtain complete coronal tree segmentation. The original 3D U-net architecture has four layers for the encoder and decoder, respectively. To improve the architecture of the 3D U-net, we added a bottleneck transformer between every two layers of the 3D U-net, and the bottleneck design uses 1×1, 3×3, and 1×1 convolution. The improved 3D U-Net architecture has 33 layers. After training, the model was validated with the validation set, and the hyperparameters were fine-tuned to optimize the model. After dealing with the distribution difference between source data and target data, different training methods of transfer learning were used to fix different model layers, adjust different learning rates for transfer learning, and fine-tune the performance by adjusting the loss function. As a result, the model of CCTA was successfully transferred to the vascular segmentation task of CMRA. The overall framework of our study is shown in Figure 1. The segmentation accuracy of coronary arteries was evaluated by dice similarity coefficient (DSC) and recall.Results
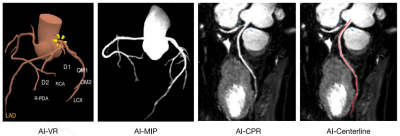
The dice similarity coefficient (DSC) of the training set and validation set were 0.944 and 0.952, and the recall were 0.923 and 0.936, respectively. The details of DSC and segmentation recall on the training and validation sets are presented in Table 1. The reconstruction results in VR, MIP, and CPR are shown in Figure 2.Discussion
Previous studies have applied deep learning to coronary vessel segmentation tasks, mainly focusing on CTA-based images. However, in clinical application, CMRA has unique advantages over CCTA. Thus, we proposed a novel and efficient automatic segmentation method for CMRA. Our study used retrospective data to train and validate the deep convolutional neural network. The results demonstrate that the proposed method can automatically obtain accurate segmentation of CMRA images. Further research and testing are needed for multi-vendor data. In future work, we will evaluate whether the image reconstruction quality meets the diagnostic needs of radiologists, and assess the efficiency of the model in clinical application.Conclusion
We present a CMRA auto-segmentation system to optimize MRA image post-processing and vessel reconstruction. It has the potential to become a standard component of daily workflows, optimizing the clinical workstation process and application of CMRA in the future.Acknowledgements
No acknowledgement found.References
1. Mortality, G. B. D. & Causes of Death, C. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study. Lancet 388(1459–1544), 2016.
2. Connolly SJ, Eikelboom JW, Bosch J, et al. Rivaroxaban with or without aspirin in patients with stable coronary artery disease: an international, randomised, double-blind, placebo-controlled trial. Lancet 2018; 391 (10117):205–218.
3. Dewey M. Coronary CT versus MR angiography: pro-CT—the role of CT angiography. Radiology. 2011 Feb;258(2):329-39.
4. Kariyasu T, Machida H, Takahashi S, et al. Denoising using deep-learning-based reconstruction for whole-heart coronary MRA with sub-millimeter isotropic resolution at 3 T: a volunteer study. Diagn Interv Radiol. 2022 Sep;28(5):470-477.
5. Hilkewich, M. W. Written observations as a part of computed tomography angiography post-processing by medical radiation technologists: a pilot project. J. Med Imaging Radiat. Sci. 45, 31–36 (2014).
6. Han D, Liu J, Sun Z, et al. Deep learning analysis in coronary computed tomographic angiography imaging for the assessment of patients with coronary artery stenosis. Comput Methods Programs Biomed. (2020) 196:105651.
7. Xu L, He Y, Luo N, et al. Diagnostic Accuracy and Generalizability of a Deep Learning-Based Fully Automated Algorithm for Coronary Artery Stenosis Detection on CCTA: A Multi-Centre Registry Study. Front Cardiovasc Med. 2021 Nov 5;8:707508.
Figures