1682
Serum Biomarkers for Risk Stratification of Prostate Cancer Patients on Active Surveillance by Untargeted 1H MRS Metabolomics1Pathology, Radiology, Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA, United States, 2Radiology, Athinoula A. Martinos Center for Biomedical Imaging, Harvard Medical School, Charlestown, MA, United States, 3Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA, United States, 4Pathology, Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA, United States, 5Urology, Massachusetts General Hospital, Boston, MA, United States
Synopsis
Keywords: Cancer, Prostate, Spectroscopy
HRMAS 1H MRS was employed for an untargeted investigation of metabolism in prostate cancer patients on AS (n=52). Serum samples from two matched patient groups, progressive (n=26) and non-progressive (n=26) were measured with short and long T2 filter to investigate metabolites in the lipoprotein and LMWM regions respectively. PCA on 46 ROI from data acquired by HRMAS 1H MRS with short T2 resulted in a metabolomic profile, PC7, that separated the progressive and non-progressive groups (p = 0.0323). The findings of such studies could revolutionize healthcare by improving the diagnosis of diseases and contributing to multi-omics integration for precision medicine.
INTRODUCTION
Prostate cancer (PCa) is a frequently diagnosed cancer and a leading cause of cancer death in men worldwide1. In recently years, PCa mortality has decreased significantly, in large part due to prostate specific antigen (PSA) screening2. This biomarker is produced in the prostate epithelium, and as a result, is prostate specific but not cancer specific. Elevation of PSA is thought to occur due to disruptions of cellular architecture and can also occur in non-malignant conditions such as benign prostatic hyperplasia (BPH) and prostatitis3.Active surveillance (AS) has emerged as the recommended strategy for patients with low-risk PCa to reduce overtreatment, as most patients will have an indolent disease course and only an estimated 20% of newly diagnosed cases are of advanced or metastatic PCa4. Several studies have successfully demonstrated the utility of ex-vivo HRMAS 1H MRS characterization of tissue in differentiating between histologically benign and malignant tissues, as well as field effects5,6. However, exploration of less-invasive biomarkers to stratify risk of tumor progression and metastasis could help improve treatment interventions and management of the disease for patients on AS7,8. The aim of this study to is to utilize untargeted metabolomics to investigate serum biomarkers in prostate cancer patients on AS that may be more sensitive and specific than the risk prediction modalities currently available in clinic.
METHODS
Patient SelectionPatients were recruited from an AS cohort of the department of urology9. For metabolomics, we identified a subpopulation that progressed in disease and received active treatment while on AS (n=26). The date of progress was defined as the date of initial active treatment triggered by Gleason grade progression, volume progression, or PSA progression. Patients who progressed in disease (P) were then matched with others from the same AS cohort who didn’t progress (N) with at 1:1 ratio (n=26). Matching was performed to exclude any confounding factors to that could affect metabolite levels based on age at diagnosis, clinical risk level (NCCN guideline), fraction of positive cores at diagnosis, Gleason score at diagnosis, PSAd, as well as the time between blood draw and prostate cancer diagnosis.
Sample Preparation
Serum samples were isolated by centrifugation from whole blood collected as a dedicated research draw. The whole blood was processed within four hours of collection and then rapidly stored in an ultralow freezer (-80°C). Samples were then transferred on dry ice to the research facility, where the blood samples were thawed at 4°C, put in solution with D2O (+TSP 0.05 wt) and a 24µl aliquot was taken. These samples were then stored at -80°C and only thawed again on ice before measurement, for a maximum of 2 freeze and thaw cycles. 12µl of blood serum was then transferred into rotors with spherical 12µl chambers for scanning.
Magnetic Resonance Spectroscopy
One-dimensional (1D) high-resolution magic angle spinning (HRMAS) proton magnetic resonance spectroscopy (1H MRS) measurements were performed on a spectrometer that consists of a 14T (600MHz) 89 mm vertical bore magnet interfaced to a Bruker Bio-Spin Avance III HD console. Data were acquired at 4˚C by CPMG sequence with water suppression and 3.6kHz spin rate. We separately performed the measurements with a short T2 filter to remove the background noise.
Data Analysis
Bruker Topspin 3.6.2 (Bruker BioSpin, Billerica, MA, USA) was utilized to process the acquired spectra. Pre-processing steps: 0.5 Hz line-broadening, one-time zero fill to 32k data points, Fourier Transform, automatic phasing and baseline correction, as well as chemical shift calibration to lactate doublet at 1.32 ppm for each spectrum. Curve-fitted peaks within the ranges of 9.5-5.1 and 4.5-0.5 ppm, purposely excluding the water region, were deconvolved. The spectral intensities for the peaks in the deconvolved regions of each spectrum were normalized by total spectral intensity. As several metabolites may contribute to a single spectral region, we identified regions of interest (ROI) with measurable spectral intensity in 50% of the individual samples for subsequent statistical analysis. 46 ROIs were identified in the data. The intensity of each ROI was normalized by the summed intensity of the ROIs in the dataset. Statistical analysis was performed in SAS JMP (Cary, NC, USA).
RESULTS and DISCUSSION
Figure 1 illustrates the results of Principal Component Analysis (PCA) on the 46 ROI. Among the calculated principal components, PC2 and PC7, as metabolomic profiles, presented higher level of significance with smaller p values. Notably, PC7 was able to separate the progressive and non-progressive groups (p = 0.0323). More detailed analyses of their effects on metabolic pathways, particularly integrations of metabolomics data with clinicopathological and single nucleotide polymorphisms (SNPs) genomic data are currently underway in our laboratory.CONCLUSION
In the future, such research efforts may enable clinicians and scientists to better understand how the pathophysiological progression of PCa is impacted by various factors present within a patient.Acknowledgements
This study is supported in part by NIH grants CA115746, CA273010, and by MGH Martinos Center for Biomedical Imaging.
References
1. Decelle EA, Cheng LL. High-Resolution Magic Angle Spinning Proton Magnetic Resonance Spectroscopy in Prostate Cancer. NMR Biomed 2014;27:10.1002/nbm.2944 doi: 10.1002/nbm.2944.
2. Manceau C, Fromont G, Beauval J-B, et al. Biomarker in Active Surveillance for Prostate Cancer: A Systematic Review. Cancers (Basel) 2021;13:4251 doi: 10.3390/cancers13174251.
3. Farha MW, Salami SS. Biomarkers for prostate cancer detection and risk stratification. Therapeutic Advances in Urology 2022;14:17562872221103988 doi: 10.1177/17562872221103988.
4. Loeb S, Bruinsma S, Nicholson J, et al. Active surveillance for prostate cancer: A systematic review of clinico-pathologic variables and biomarkers for risk stratification. Eur Urol 2015;67:619–626 doi: 10.1016/j.eururo.2014.10.010.
5. Steiner A, Schmidt SA, Fellmann CS, et al. Ex Vivo High-Resolution Magic Angle Spinning (HRMAS) 1H NMR Spectroscopy for Early Prostate Cancer Detection. Cancers (Basel) 2022;14:2162 doi: 10.3390/cancers14092162.
6.Dinges SS, Vandergrift LA, Wu S, et al. Metabolomic Prostate Cancer Fields in HRMAS MRS-profiled Histologically Benign Tissue Vary with Cancer Status and Distance to Cancer. NMR Biomed 2019;32:e4038 doi: 10.1002/nbm.4038.
7. Bar N, Korem T, Weissbrod O, et al. A reference map of potential determinants for the human serum metabolome. Nature 2020;588:135–140 doi: 10.1038/s41586-020-2896-2.
8. Psychogios N, Hau DD, Peng J, et al. The Human Serum Metabolome. PLOS ONE 2011;6:e16957 doi: 10.1371/journal.pone.0016957.
9. Salari K, Kuppermann D, Preston MA, et al. Active Surveillance of Prostate Cancer is a Viable Option for Men Younger than 60 Years. J Urol 2019;201:721–727 doi: 10.1097/JU.0000000000000031.
Figures
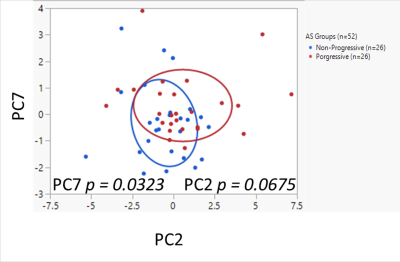
Figure 1. Principal Component Analysis (PCA) results on 46 ROI from data acquired by HRMAS 1H MRS with short T2 to investigate lipoproteins (LP) in the 3.3-0.5 ppm region. Non-progressive group (n=26) labeled in blue color, progressive group (n=26) in red. The bivariate normal ellipses represent p = 0.50.