1669
Tumor segmentation with nnU-Net on dynamic contrast enhanced MR images of triple negative breast cancer
Zhan Xu1, Sanaz Pashapoor2, Bikash Panthi1, Jong Bum Son1, Ken-Pin Hwang1, Beatriz Elena Adrada2, Rosalind Pitpitan Candelaria2, Mary Saber Guirguis2, Miral Mahesh Patel2, Huong Le-Petross2, Jessica Leung2, Marion Elizabeth Scoggins2, Gary Whitman2, Rania Mohamed2, Deanna Lynn Lane2, Tanya Moseley2, Frances Perez2, Jason White3, Elizabeth Ravenberg3, Alyson Clayborn3, Huiqin Chen4, Jia Sun4, Peng Wei4, Alastair Thompson5, Anil Korkut6, Lei Huo7, Kelly Hunt8, Stacy Moulder3, Jennifer Litton3, Vicente Valero3, Debu Tripathy9, Wei Yang2, Clinton Yam3, Gaiane Margishvili Rauch2, and Jingfei Ma1
1Department of Imaging Physics, MD Anderson Cancer Center, Houston, TX, United States, 2Departments of Breast Imaging, MD Anderson Cancer Center, Houston, TX, United States, 3Departments of Breast Medical Oncology, MD Anderson Cancer Center, Houston, TX, United States, 4Departments of Biostatistics, MD Anderson Cancer Center, Houston, TX, United States, 5Section of Breast Surgery, Baylor College of Medicine, Houston, TX, United States, 6Departments of Bioinformatics & Computational Biology, MD Anderson Cancer Center, Houston, TX, United States, 7Departments of Pathology, MD Anderson Cancer Center, Houston, TX, United States, 8Departments of Breast Surgical Oncology, MD Anderson Cancer Center, Houston, TX, United States, 9Department of Breast Medical Oncology, MD Anderson Cancer Center, Houston, TX, United States
1Department of Imaging Physics, MD Anderson Cancer Center, Houston, TX, United States, 2Departments of Breast Imaging, MD Anderson Cancer Center, Houston, TX, United States, 3Departments of Breast Medical Oncology, MD Anderson Cancer Center, Houston, TX, United States, 4Departments of Biostatistics, MD Anderson Cancer Center, Houston, TX, United States, 5Section of Breast Surgery, Baylor College of Medicine, Houston, TX, United States, 6Departments of Bioinformatics & Computational Biology, MD Anderson Cancer Center, Houston, TX, United States, 7Departments of Pathology, MD Anderson Cancer Center, Houston, TX, United States, 8Departments of Breast Surgical Oncology, MD Anderson Cancer Center, Houston, TX, United States, 9Department of Breast Medical Oncology, MD Anderson Cancer Center, Houston, TX, United States
Synopsis
Keywords: Cancer, Breast, TNBC
Quantitative image analysis of cancers requires accurate tumor segmentation that is often performed manually. In this study, we developed a deep learning model with the self-configurable nnU-Net for automated tumor segmentation on dynamic contrast enhanced MR images of triple negative breast cancers. Our results on an independent testing dataset demonstrated that this nnU-Net based deep learning model can perform automated tumor segmentation with high sensitivity and Dice coefficient.Introduction:
Triple-negative breast cancer (TNBC) is a biologically aggressive and heterogenous subtype of breast cancer that is treated with neoadjuvant systemic therapy but has varied responses, which predict long-term outcome. Quantitative imaging has been investigated and shown potential to predict the early1. As an important step in the quantitative image analyses, tumor segmentation is usually performed by specially trained personnel in a manual and often laborious manner. Manual tumor segmentation is time-consuming and susceptible to inter- and intra- reader variability. In recent years, deep learning (DL) models have shown great success and potential for automated biomedical image segmentation applications2-4. In particular, nnU-Net5 is a self-configurable DL framework and its segmentation performance is comparable or even surpasses many customized DL models in terms of Dice coefficient (DICE). In this study, we developed a DL model by using nnU-Net on dynamic contrast-enhanced (DCE)-MR images of TNBC patients acquired at baseline before the initiation of therapy and we evaluated the model segmentation performance when different images were used as the model inputs.Method:
Datasets: We included 162 biopsy-confirmed stage I-III TNBC patients in our study. All the patients were enrolled in an IRB approved prospective clinical trial (NCT02276443) and underwent baseline DCE MRI scan; 108 and 27 patients were used for training and validation, and another 27 patients for testing. DCE images were acquired using a 3D T1-weighted DISCO sequence axially and with the following parameters: acquisition matrix: 320×320, field-of-view:300×300mm, number of slice: 112~192, slice thickness: 3.2mm, slice gap: -1.6mm, TR/TE=3.9/1.7ms, flip angle= 12°, number of phases: 45~62, temporal resolution: ~10s. The segmentation of tumor ROIs from early phase (60s) DCE subtraction images was carried out by two fellowship-trained breast radiologists. The manually segmented tumor ROIs were used as the reference standards for the model development and testing. All DCE images and masks were zero-padded to 192 slices before model training.Model Training: We trained 3D nnU-Net with default hyperparameter setups that were provided by the model developers as the public download of the source code5. Five-fold cross-validation was used on training/validation sets. Five different combinations of raw DCE images and semi-quantitative parametric maps were used as the model inputs: 1. DIFF: difference images between the DCE images at 60s after contrast agent injection and the pre-injection images, 2. PEI: positive enhancement integral images, 3. SER: signal enhancement ratio images, 4. MSI: maximum slope of increase images, 5. COMB: the combination of DIFF, PEI, and MSI images. The PEI, SER, and MSI images were calculated using the MR scanner vendor provided software. The first 4 inputs were trained in 3D mode and COMB was trained in 4D mode by which DIFF, PEI and MSI images were concatenated as the 4th dimension.
Model Testing: Segmentation performance was evaluated on the independent testing dataset with DICEs, and sensitivity for each subject, then summarized over the testing set6.
Result & Discussion:
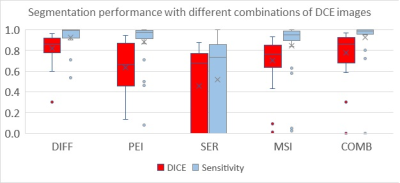
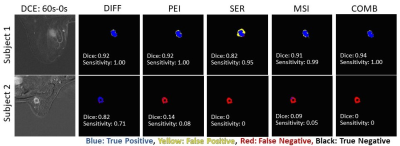
Models with DIFF, PEI, MSI, and COMB as inputs yielded good segmentation performance with DICEs, and sensitivity > 0.8 (Fig. 1). The model performance in one subject is shown in Fig. 2 (top row). In comparison, the model with SER as input performed poorly, with group DICE/sensitivity = 0.46/0.52. The model with DIFF as input yielded the best segmentation performance with a DICE of 0.82 and sensitivity of 0.92, which is slightly lower than that of COMB (sensitivity of 0.93). In one subject, the model with COMB failed to identify the tumor (sensitivity of 0) while the model with DIFF yielded a sensitivity of 0.71 (subject 2 in the bottom row of Fig. 2). Upon DCE image examination, this subject had a large artifact from the biopsy clip at the center of the tumor, which led to an excluded hollow area within the tumor in the manually contoured tumor ROI. It is conceivable that the current model requires additional cases for training to further improve the model performance.Conclusion:
We have successfully developed a DL model for automated tumor segmentation using nnU-Net on DCE MR images of TNBC. Our investigation showed that different combinations of the DCE images and semi-quantitative parametric maps can be used as inputs to the model. An optimal model may be established with a larger number of patients for training and testing. Such a model will be highly useful for quantitative image analyses in a variety of clinical applications.Acknowledgements
No acknowledgement found.References
- Uematsu T. MR imaging of triple-negative breast cancer. Breast Cancer. Jul 2011;18(3):161-4.
- Alom MZ, Yakopcic C, Hasan M, Taha TM, Asari VK. Recurrent residual U-Net for medical image segmentation. J Med Imaging (Bellingham). Jan 2019;6(1):014006.
- Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Springer International Publishing; 2015:234-241.
- He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2016:770-778.
- Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods. Feb 2021;18(2):203-211.
- Taha AA, Hanbury A. Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med Imaging. Aug 12 2015;15:29.
DOI: https://doi.org/10.58530/2023/1669