1667
Habitat Analysis Based on Multiparametric MRI Predicts Lung Adenocarcinoma Subtypes1Department of Radiology, Fourth Hospital of Hebei Medical University, Shijiazhuang, China, 2Central Research Institute, United Imaging Healthcare, Shanghai, China
Synopsis
Keywords: Cancer, Lung
As a promising approach to analyze tumor heterogeneity through image features, habitat analysis has been applied for a variety of tumors, such as lung adenocarcinoma (LUAD). To our knowledge, the previous applications, mainly based on CT and PET images, would potentially hold a limited impact on the power of habitats analysis. In this study, our goal is to extend the applications of habitat analysis in LUAD with multi-parametric MR images, which would be obtained routinely longitudinally and to guide specific therapies.Introduction
LUAD is the most common type in non–small cell lung cancer (NSCLC) with a 5-year survival rate lower than 20%.1 World Health Organization (WHO) reported that most LUAD has a mixed-subtype, and the degree of the histopathologic diversity within the tumor is highly associated with patient prognosis. Mounting evidence demonstrated that various subtypes are largely held to be responsible for the aggressive progression and inferior responses to anticancer therapies.2, 3 Therefore, accurate histological subtype classification is vital for therapeutic decision-making.4.5 Biopsy is the gold standard for the classification, but the limited area was unable to represent the entire tumor. To date, a novel method named habitat analysis has been proposed to evaluate the tumor physiology information and its changes with growth or the response to therapy in real-time and long-term. For lung cancer, most studies using habitat analysis were developed based on CT and PET, of which the radiation exposure is still a concern. As such, the noninvasive multi-parametric MRI is a better choice for habitat analysis, not only for its radiation free property but for the functional information it could provide. The aim of the study is to investigate the clinical feasibility of tumor habitat analysis based on multi-parametric MRI in differentiating predominant subtypes of LUAD.Methods
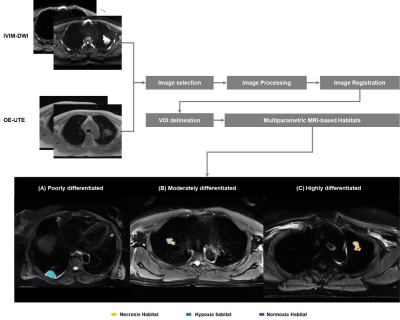
Patients with confirmed LUAD were evaluated on 3T MR scanner (uMR780, United Imaging Healthcare, Shanghai, China) using intravoxel incoherent motion diffusion-weighted imaging (IVIM-DWI) (b = 0, 10, 20, 30, 50, 80, 100, 200, 400 and 800 s/mm2) and oxygen-enhanced MRI with ultrashort echo time (OE-UTE). The quantitative parameters from IVIM (D, D* and f) were calculated using a post-processing workstation (United Imaging Healthcare, Shanghai, China). The voxel-wise percent signal enhancement (PSE) map from OE-MRI of each patient was obtained with the previous algorithm.6 Primary tumor volumes of interest (VOIs) were labeled by a radiologist with 10-years’ experience and reviewed by a senior radiologist with 20-years’ experience. K-means clustering algorithm in Scikit-Learn Python package was used for classifying different tumor habitats based on D maps and PSE maps. Then, three intratumor habitats were identified: the necrosis area with high D value and low PSE habitat, the hypoxia tumor subregion with low D value and low PSE, and the normoxia tumor subregion with high PSE, respectively. Subsequently, several quantitative parameters including tumor volume and volume fraction were calculated on each patient’s corresponding tumor subregions. Habitats parameters were expressed as mean value with standard deviation (SD). Kruskal-Wallis One-Way ANOVA test was performed to compare the fraction of each cluster among the three subtypes. The classification performance was evaluated using the area under the receiver operating characteristic curve (AUC).Results
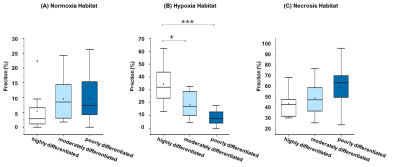
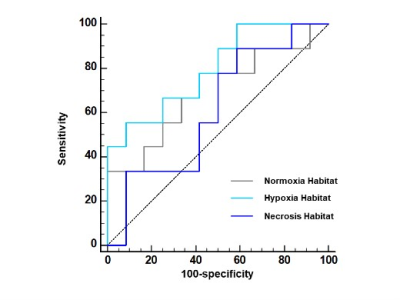
A total of thirty-three patients were recruited in this study. The number of highly differentiated (adenocarcinoma in situ, minimally invasive adenocarcinoma or lepidic predominant adenocarcinoma), moderately differentiated (papillary or acinar predominant adenocarcinoma) and poorly differentiated (micropapillary, or solid predominant adenocarcinoma) subtypes was 12, 12, and 9, respectively. Figure 1 showed the workflow and examples of clustering maps. A relatively large difference in the hypoxia habitat between three subtypes was observed, which indicated successful discrimination. The distributions of volume fraction for each habitat were presented in Figure 2. The poorly differentiated group had a higher volume fraction in hypoxia habitat than the moderately and highly differentiated groups (P < 0.05). Moreover, the diagnostic model using the volume fraction of hypoxia habitat (AUC = 0.796) yielded better discriminatory ability for differentiating tumor subtypes of LUAD than the necrosis habitat (AUC = 0.611) and normoxia tumor habitat (AUC = 0.694), figure 3.Discussion
In this study, an unsupervised data-driven clustering method based on D and PSE maps derived from multi-parametric MRI was proposed to split the tumor volume into three functional habitats. The volume fraction in hypoxia habitat provided independent prognostic value beyond this in other habitats to stratify patients into different subtypes. Compared to CT and PET, MRI allows a multidimensional in vivo characterization of lung cancer, including structural, physiologic, and functional information without the risk of radiation exposure. The habitat analysis based on multi-parametric MRI in lung cancer has been firstly evaluated. Additionally, hypoxia is the key determinant of tumor subregions towards aggressiveness, resistance to therapy and poor patient outcomes.7 Higher percentages of hypoxic tumor regions were predictive of metastasis development in patients, which is in lines with the results of this study. This study has several limitations. The main limitation was the limited number of patients. Besides, a particular protocol for OE-MRI and IVIM imaging was used in this study which is not universal in clinical practice.Conclusion
In conclusion, the volume fraction of hypoxia habitat based on multi-parametric MRI could be a promising parameter in LUAD histological subtypes classification.Acknowledgements
None.References
1.Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2020. CA Cancer J Clin 70, 7-30, doi:10.3322/caac.21590 (2020).
2.Dagogo-Jack, I. & Shaw, A. T. Tumour heterogeneity and resistance to cancer therapies. Nature Reviews Clinical Oncology 15, 81-94, doi:10.1038/nrclinonc.2017.166 (2017).
3.Vitale, I., Shema, E., Loi, S. & Galluzzi, L. Intratumoral heterogeneity in cancer progression and response to immunotherapy. Nat Med 27, 212-224, doi:10.1038/s41591-021-01233-9 (2021).
4.Travis, W. D. et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 6, 244-285, doi:10.1097/JTO.0b013e318206a221 (2011).
5.Warth, A. et al. The novel histologic International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification system of lung adenocarcinoma is a stage-independent predictor of survival. J Clin Oncol 30, 1438-1446, doi:10.1200/JCO.2011.37.2185 (2012).
6.Liu, H. et al. Pulmonary Functional Imaging for Lung Adenocarcinoma: Combined MRI Assessment Based on IVIM-DWI and OE-UTE-MRI. Front Oncol 11, 677942, doi:10.3389/fonc.2021.677942 (2021).
7.Freeman, S. J. et al. The revised FIGO staging system for uterine malignancies: implications for MR imaging. Radiographics 32, 1805-1827, doi:10.1148/rg.326125519 (2012).
Figures