1652
Using convolutional neural network predicts microvascular invasion in hepatocellular carcinoma based on Gd-EOB-DTPA-enhanced MRI1Nanfang Hospital, Southern Medical University, Guangzhou, China, 2Guangzhou University of Chinese Medicine, Guangzhou, China
Synopsis
Keywords: Liver, Cancer, Gd-EOB-DTPA-enhanced MRI
An accurate preoperative assessment of microvascular invasion (MVI) in patients with hepatocellular carcinoma (HCC) is of great clinical importance in choosing appropriate surgical interventions. We aimed to investigate diagnostic performance of Gd-EOB-DTPA-enhanced MRI for prediction of MVI in HCC using convolutional neural network (CNN). The CNN model based on hepatobiliary phase (HBP) images had great diagnostic efficiency for the prediction of MVI with the AUC of 0.858 (range, 0.854, 0.893). Deep learning with CNN based on Gd-EOB-DTPA-enhanced MRI can be conducive to preoperative prediction of MVI in HCC.Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignancies and has become the third leading cause of cancer-related deaths worldwide1. Microvascular invasion (MVI) in HCC is considered to be a critical determinant of recurrence after surgical resection and liver transplantation2. Therefore, preoperative knowledge of MVI is of great importance in selecting appropriate treatment strategies. Deep learning has been shown superiority in feature representation for lesion characterization3,4. To the best of our knowledge, very few studies have applied deep learning approach based on Gd-EOB-DTPA-enhanced MRI to predict MVI in HCC5. Therefore, the purpose of this study was to develop and evaluate a deep learning model based on hepatobiliary phase (HBP) images of Gd-EOB-DTPA-enhanced MRI to accurately predict preoperative MVI in HCC.Methods
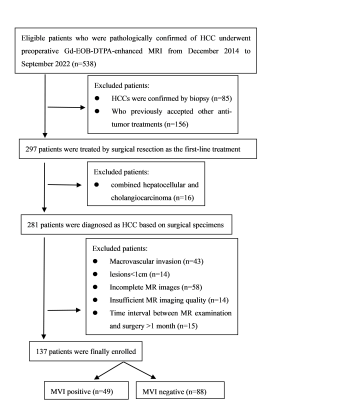
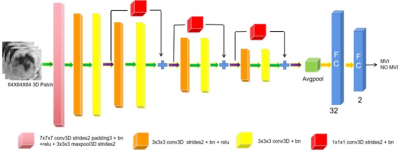
This retrospective study included 137 patients with pathologically confirmed HCC who underwent Gd-EOB-DTPA-enhanced MRI before surgery from December 2014 to September 2022. Patients were randomly divided into a training cohort (n=96; MVI-positive, n=35; MVI-negative, n=61) and a validation cohort (n=41; MVI-positive, n=14; MVI-negative n=27) in a ratio of 7:3. Flow chart of patients’ recruitment is shown in Figure 1. All MRI examinations were performed by a 3.0 T system (Achieva, Philips Healthcare, The Netherlands). For dynamic MRI, gadoxetic acid (Primovist, Bayer Schering Pharma, Berlin, Germany) with a concentration of 0.025 mmol/kg was injected at a flow rate of 2.0 mL/s, followed by a 20-mL saline flush. A fat-suppressed three-dimensional sequence (TR/TE = 3.1 ms/1.51 ms, 304 × 239 matrix, 5-mm slice thickness) was performed before injection of the contrast agent. Arterial phase (15-20 s), portal venous phase (40-60 s), delayed phase (120-180 s) and hepatobiliary phase (20 min) were respectively obtained after the injection of agent using the same sequences as the pre-contrast images. Demographic and clinicopathological variables of all patients were retrieved from medical record system and pathological reports. MR imaging features were analyses independently by two radiologists. In case of any discrepancy, a consensus was reached after discussion with a senior radiologist. The volumes of interest (VOIs) were delineated around the HCC lesions outline for 3D volume area as indicated in HBP images by a radiologist independently with ITK-Snap software (http://www.itksnap.org), and then confirmed by a senior radiologist. Then, we extracted volumetric tumor areas from original images using MATLAB (The MathWorks, Inc.). Data augmentation used the image resampling method to generate more 3D samples from the current limited tumor area to train the deep learning network. In this work, we used the 3D ResNet model to predict the MVI status in HCC based on HBP images. In detail, the framework of the proposed deep learning model is shown in Figure 2. Two independent sample t-test, Mann–Whitney U test, chi-square test or Fisher exact test were used to compare clinical characteristics and radiological features between the MVI-positive and MVI-negative groups in the training cohort and validation cohort, as appropriate. Receiver operating characteristic curve (ROC) analysis was used to evaluate the performance for MVI prediction in the validation cohort. A two-tailed p < 0.05 was considered statistically significant. All statistical analyses were performed using IBM SPSS statistical software (version 26).Results
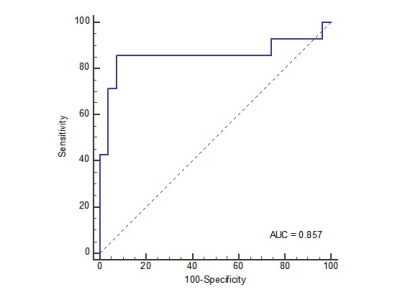
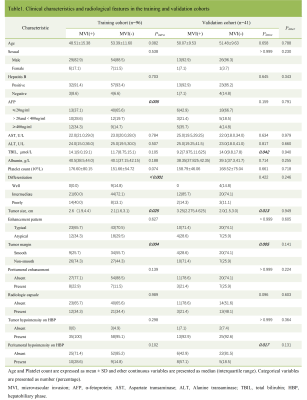
The clinical characteristics and radiological features in the training and validation cohorts are listed in Table 1. It could be found that only the tumor size and tumor margin had statistical difference to differentiate the MVI status in both training and validation cohorts. The diagnostic performance of the tumor size and tumor margin was evaluated in the validation cohort with cutoff values determined in the training cohort. The AUC, sensitivity, specificity and accuracy of the tumor size and tumor margin were 0.706 (95% CI 0.543-0.870), 85.7%, 55.6%, 65.9% and 0.728 (95% CI, 0.559-0.896), 71.4%, 74.1%, 73.2%, respectively. The indicators of performance evaluation of the deep learning model, including AUC, sensitivity, specificity and accuracy were expressed as median (range, minimum, and maximum) of five repeated measurements in the validation cohort. The AUC, sensitivity, specificity and accuracy of the deep learning model based on HBP images were 0.858 (range, 0.854, 0.893), 0.743 (range, 0.690, 0.764), 0.859 (range, 0.832, 0.915), 0.820 (range, 0.783, 0.867). Figure 3 shows the ROC curve of the deep learning model.Discussion
In this study, we proposed a CNN based on HBP images for MVI prediction in HCC. The reasons that the CNN model achieved high performance might be explained as follows. First, Gd-EOB-DTPA-enhanced MRI can accurately discriminate lesion boundaries. The difference in signal between tumor tissues and surrounding normal liver parenchyma is more obvious in the HBP images, which makes the boundaries of tumors clearer to delineate. Then, deep features obtained using CNN from data-driven learning have proven the superior performance for lesion characterization in medical imaging processing. In addition, we have applied the 3D CNN to predict MVI which can make use of the three-dimensional spatial information in volumetric data to more accurately characterize the lesions. Our future work will consider the fusion of clinical characteristics and radiological features in the CNN for performance improvement.Conclusion
Our study suggested that a deep learning model based on HBP images of Gd-EOB-DTPA-enhanced MRI achieved high diagnostic performance for MVI prediction in HCC preoperatively.Acknowledgements
The authors thank the School of Medical Information Engineering, Guangzhou University of Chinese Medicine for providing technical support for this study.References
1. Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nature reviews Disease primers. Jan 21 2021;7(1):6. doi:10.1038/s41572-020-00240-3
2. Erstad DJ, Tanabe KK. Prognostic and Therapeutic Implications of Microvascular Invasion in Hepatocellular Carcinoma. Annals of surgical oncology. May 2019;26(5):1474-1493. doi:10.1245/s10434-019-07227-9
3. Hamm CA, Wang CJ, Savic LJ, et al. Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi-phasic MRI. European radiology. Jul 2019;29(7):3338-3347. doi:10.1007/s00330-019-06205-9
4. Wang G, Jian W, Cen X, et al. Prediction of Microvascular Invasion of Hepatocellular Carcinoma Based on Preoperative Diffusion-Weighted MR Using Deep Learning. Academic radiology. Dec 7 2020;doi:10.1016/j.acra.2020.11.014
5. Wei J, Jiang H, Zeng M, et al. Prediction of Microvascular Invasion in Hepatocellular Carcinoma via Deep Learning: A Multi-Center and Prospective Validation Study. Cancers (Basel). May 14 2021;13(10)doi:10.3390/cancers13102368
Figures