1650
Multiparametric MR-based Feature Fusion Radiomics Combined with ADC Maps-based Tumor Proliferative Burden in Distinguishing TNBC vs. non-TNBC
Fangrong Liang1,2, Wanli Zhang1,2, Jiamin Li1,2, Yongzhou Xu3, Aaron Zhang3, Xinqing Jiang1,2, Xin Zhen4, and Ruimeng Yang1,2
1Department of Radiology, The Second Affifiliated Hospital, School of Medicine, South China University of Technology, Guangzhou, China, 2Department of Radiology, Guangzhou First People’s Hospital, Guangzhou, China, 3Philips Healthcare, Guangzhou, China, 4School of Biomedical Engineering, Southern Medical University, Guangzhou, China
1Department of Radiology, The Second Affifiliated Hospital, School of Medicine, South China University of Technology, Guangzhou, China, 2Department of Radiology, Guangzhou First People’s Hospital, Guangzhou, China, 3Philips Healthcare, Guangzhou, China, 4School of Biomedical Engineering, Southern Medical University, Guangzhou, China
Synopsis
Keywords: Breast, Diffusion/other diffusion imaging techniques, Tumor proliferative burden; Triple negative breast cancer; Multiparametric magnetic resonance imaging
Triple negative breast cancer (TNBC) is highly heterogeneous, with poorer prognosis, higher recurrence rates and severe treatment challenges. Accurate preoperative identification of TNBC is helpful for individualized patient management. Based on multiparametric magnetic resonance imaging (mMRI), we employed whole-tumor ADC maps-based radiomics (RADC) model, tumor proliferative burden (TPBADC) model, mMRI-based feature fusion radiomics (RFF) model and combinational RFF-TPBADC model to investigate their performance in distinguishing TNBC from non-TNBC. Our results showed that the RFF-TPBADC model outperformed the RADC, TPBADC, and RFF models by integrating mMRI radiomics features and TPBs, demonstrating its potential for classification of breast cancer.Introduction
Previous studies have demonstrated that ADC maps could be applied to preoperative identification of triple negative breast cancer (TNBC) by generating quantitative ADC cutoffs (e.g., minimum, mean, or maximum ADC)1, 2. It is well known that absolute ADC values are susceptible to various b-values from different MR scanners and different medical centers3. Tumor proliferative burden (TPB), which was defined as the proportion of the sum of the lower ADC values in the tumor’s overall pixel set, has been successfully used to assess the prognosis of metastatic renal cell carcinoma patients treated with neoadjuvant sunitinib4. However, the contribution of TPB to TNBC identification has not been fully established. Therefore, based on a newly lab-developed mMRI-based feature fusion radiomics (RFF) model, our study aimed to investigate whether the addition of TPB could help improve the distinguishing performance of TNBC in an mMRI-based RFF radiomics model.Methods
Data collectionThis retrospective study included 460 patients with pathologically proven BCs (TNBC: n = 54 vs. non-TNBC: n = 412) who underwent breast MRI examinations on 1.5T (uMR 560, United Imaging) between January 2017 to April 2022 in Guangzhou First People’s Hospital (Figure 1). The MRI sequences included T1WI, T2WI, DWI with b values of b=0 s/mm2, 600 s/mm2, and six phases of dynamic contrast-enhanced MRI (DCEphase1-6) with approximately 62s per acquisition. ADC maps were automatically generated on the post-processing workstation. All cases were randomly divided into the training set (n=337) and test set (n=129) at a ratio of 1:3.
Tumor segmentation and radiomics feature selection
The volume of interest (VOI) of each lesion was delineated slice-by-slice on T2WI, DWI, ADC maps and DCEphase2 images. By using Pyradiomics, an open-source software toolkit for radiomics analysis5, a total of 109 radiomics features were extracted from each VOI, which were then fed into 150 classification models constructed with 10 classifiers and 15 feature selection methods.
Model development and evaluation
The following four models were developed (Figure 2):
i. Whole-tumor ADC maps-based radiomics (RADC) model: based on radiomics features extracted from ADC maps.
ii. TPBADC model:
1) A pixel-based ADC value histogram of the whole tumor on the ADC map was obtained;
2) The top 1-25% lower ADCs (denoted as ADC1st, ADC2nd, ... ADC24th, ADC25th), the mean and median ADCs (denoted as ADCmean and ADCmedian), the mean of the top 25% and 75% lower ADCs (denoted as ADCtop25% mean and ADCtop75% mean), the top 1-25% lower TPBs (denoted as TPB1st, TPB2ed, ...TPB24th, TPB25th), and four groups of TPBs (denoted as TPB≤5th, TPB≤10th, TPB≤20th and TPB≤25th) were calculated based on the ADC histogram.
iii. RFF model: fusion of multi-sequences (T2WI, DWI, ADC maps and DCEphase2) radiomics feature.
iv. RFF-TPBADC model: integrating the RFF model and the TPBADC model.
The performances of the four models were compared via the area under the receiver operative characteristic curve (AUC), sensitivity (SEN), specificity (SPE) and accuracy (ACC).
Results
Both the RFF model and TPBADC model demonstrated good performance in distinguishing TNBC from non-TNBC, with a maximum AUC value of 0.818, 0.808 and 0.787, 0.718 in the training set and test set, respectively. In the TPBADC model, all the top 10 most discriminative parameters were TPB-related measures other than ADC cutoffs as shown in Figure 3. Compared with the non-TNBC group, the top 1-25% lower ADCs in the TNBC group were lower than those of non-TNBC (Figure 4). Integrating the most discriminative features, By combining the RFF model and TPBADC model, the RFF-TPBADC model displayed superior efficiency to the RADC, RFF, and TPBADC model, with a higher AUC of 0.839 and 0.791 in the training set and test set, respectively (p < 0.05) (Table 1).Discussion
ADCs directly reflect tumor cellularity, water content and cell membrane integrity in the tumor microenvironment6. Interestingly, our results showed that other than ADC cutoffs, all of the top 10 most discriminative parameters of the TPBADC model were TPB-related measurements. The top 1-25% lower ADCs were lower in the TNBC group than those in the non-TNBC group, which represents the most restricted cellularity of the tumor, reflecting the more malignant biological characteristics of TNBC. Avoiding the drawbacks of absolute ADCs that are susceptible to different MR scanners and various b values under different scan protocols3, our study employed TPB-related measurements and found that TPB has a good performance in distinguishing TNBC from non-TNBC. Recently, a novel RFF model was successfully developed in our lab based on a hypothesis that managing MRI sequences in a collaborative pattern would provide multi-dimensional image information by incorporating class structure information7. Our results also demonstrated that features of the RFF model fused with four sequences outperformed the RADC model (p<0.05). Notably, by combining the TPBADC model with the RFF model, the new RFF-TPBADC model outperformed any other model to distinguish TNBC from non-TNBC, which indicated TPBADC could contribute to improving the performance of the RFF model.Conclusion
We developed an mMR-Based RFF model cooperating with a whole-tumor ADC maps-based tumor proliferative burden model (RFF-TPBADC model) to identify TNBC and non-TNBC preoperatively. The RFF-TPBADC model is an effective and reliable tool that has great potential in tumor diagnosis or prognostic prediction.Acknowledgements
No acknowledgement found.References
1. Horvat J V, Bernard-Davila B, Helbich T H, et al. Diffusion-weighted imaging (DWI) with apparent diffusion coefficient (ADC) mapping as a quantitative imaging biomarker for prediction of immunohistochemical receptor status, proliferation rate, and molecular subtypes of breast cancer. J Magn Reson Imaging, 2019,50(3):836-846.2. Xie T, Zhao Q, Fu C, et al. Differentiation of triple-negative breast cancer from other subtypes through whole-tumor histogram analysis on multiparametric MR imaging. European Radiology, 2019,29(5):2535-2544.
3. Marino M A, Avendano D, Sevilimedu V, et al. Limited value of multiparametric MRI with dynamic contrast-enhanced and diffusion-weighted imaging in non-mass enhancing breast tumors. Eur J Radiol, 2022,156:110523.
4. BHARWANI N, MIQUEL M E, POWLES T, et al. Diffusion-weighted and multiphase contrast-enhanced MRI as surrogate markers of response to neoadjuvant sunitinib in metastatic renal cell carcinoma. British journal of cancer, 2014,110(3):616-624.
5. Haghighat M, Abdel-Mottaleb M, Alhalabi W. Discriminant Correlation Analysis: Real-Time Feature Level Fusion for Multimodal Biometric Recognition. IEEE transactions on information forensics and security, 2016,11(9):1984-1996.
6. Koh D M, Collins D J. Diffusion-weighted MRI in the body: applications and challenges in oncology. AJR Am J Roentgenol, 2007,188(6):1622-1635.
7. Wu J, Liang F, Wei R, et al. A Multiparametric MR-Based RadioFusionOmics Model with Robust Capabilities of Differentiating Glioblastoma Multiforme from Solitary Brain Metastasis. Cancers (Basel), 2021,13(22).
Figures
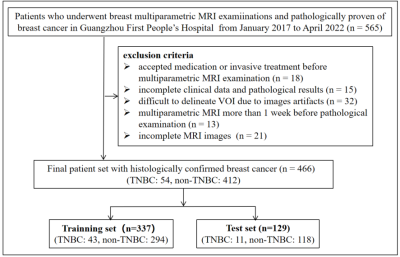
Figure 1. Flow chart of the study population with inclusion and exclusion criteria. TNBC, triple negative breast cancer.
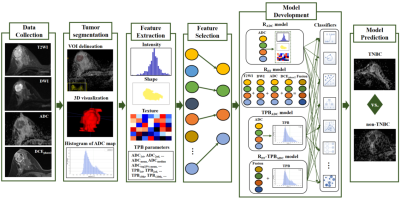
Figure 2. Flow chart of this study. TNBC, triple negative breast cancer.
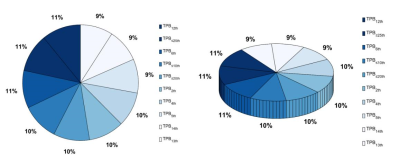
Figure 3. A pie charts showing the number of times (%) that the TPBADC model was selected to include the top 10 most discriminative parameters.
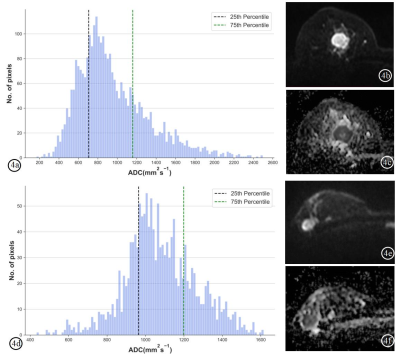
Figure 4. Whole-tumor apparent diffusion coefficient (ADC) maps-based histogram, DWI (b=600 s/mm2) and ADC images of a TNBC patient (4a-4c) and a non-TNBC patient (4d-4f). The top lower ADC25th in the TNBC patient was lower than that of non-TNBC patient (706 s/mm2 vs. 964 s/mm2). ADC, apparent diffusion coefficient. TNBC: triple-negative breast cancer.
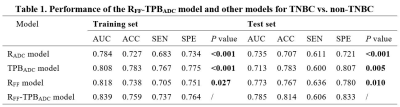
Table 1. performance of the RFF-TPBADC model and other models for TNBC vs. non-TNBC.
P value: compared the performance between the RFF-TPBADC model and other models via paired samples Wilcoxon signed rank test in the training set and test set. Significant values (p < 0.05) are presented in bold. AUC, area under the receiver-operating characteristic curve; SEN, sensitivity; SPE, specificity; ACC, accuracy.
DOI: https://doi.org/10.58530/2023/1650