1648
AI-based reconstruction of T2-weighted sequences in prostate MRI: clinical evaluation and impact on diagnostic confidence
Leon Bischoff1,2, Alexander Isaak1,2, Christoph Katemann3, Dmitrij Kravchenko1,2, Narine Mesropyan1,2, Christoph Endler1,2, Barbara Wichtmann1,2, Oliver Weber3, Johannes Peeters4, Claus Christian Pieper1, Daniel Kütting1,2, Alois Martin Sprinkart1,2, Ulrike Attenberger1, and Julian Luetkens1,2
1Department of Diagnostic and Interventional Radiology, University Hospital Bonn, Bonn, Germany, 2Quantitative Imaging Lab, University Hospital Bonn, Bonn, Germany, 3Philips GmbH Market DACH, Hamburg, Germany, 4Philips MR Clinical Science, Best, Netherlands
1Department of Diagnostic and Interventional Radiology, University Hospital Bonn, Bonn, Germany, 2Quantitative Imaging Lab, University Hospital Bonn, Bonn, Germany, 3Philips GmbH Market DACH, Hamburg, Germany, 4Philips MR Clinical Science, Best, Netherlands
Synopsis
Keywords: Prostate, Machine Learning/Artificial Intelligence, Prostate cancer
In this prospective study, 56 male patients with suspected prostate cancer were included to evaluate an artificial intelligence (AI) based reconstruction method for T2-weighted sequences in multiparametric MRI (mpMRI). After comparison with conventionally acquired and reconstructed sequences we found the new AI-based method to produce images with higher image sharpness and delineation of lesions, confirmed by both qualitative and quantitative analysis. This is accompanied by a reduction of scan time by 29-37%. Confidence in the assessed PI-RADS scores was significantly higher for the AI-reconstruction. This new technique could therefore potentially increase diagnostic accuracy of mpMRI of the prostate.Introduction
With an aging population fast and efficient MRI scans are needed to satisfy the increasing demand. Especially prostate cancer ranks among the most prevalent cancer types in men1. An early and precise diagnosis by multiparametric MRI (mpMRI) is therefore not only crucial for long-term survival but also lays the foundation for the invasive procedure of a targeted biopsy2. In recent years artificial intelligence (AI) has become one of the most promising methods in medicine; the field of radiology benefits greatly from these advances due to the high potential for image analysis3. The aim of this study was therefore to investigate qualitatively and quantitatively AI-enhanced T2-sequences in the setting of prostate mpMRI and whether it results in a better diagnostic confidence using the PI-RADS classification4.Material and Methods
The prospective study was approved by the local review committee and all subjects gave written consent prior to mpMRI. Inclusion criteria were either an elevated prostate-specific-antigen of >4ng/ml or a suspicious digital rectal exam/transrectal ultrasound. All scans were performed on a 3 Tesla MRI (Philips Ingenia 3T). Besides T1- and diffusion-weighted sequences the scan protocol included a T2-weighted (T2w) cartesian low-resolution (T2LR), a T2w cartesian high-resolution (T2HR) and a T2w propeller (T2PR) sequence. Derived from the raw data of the LR-sequence one additional sequence (T2AI) was reconstructed using vendor provided prototype (Philips SmartSpeed Precise Image, SSPI). This AI-based reconstruction technique consists of a series of convolutional neural networks (CNNs): Adaptive-CS-Net5 allows to reconstruct images acquired with Compressed SENSE based variable density undersampling patterns. This CNN is applied prior to coil combination, removing the noise and undersampling artifacts from the images to obtain good image quality from accelerated acquisitions6. Subsequently, SSPI is an AI-model applied to replace the traditional zero-filling strategy and to increase the matrix size and therewith the sharpness of the images (super resolution)7,8. This network is trained on pairs of low- and high-resolution data with k-space crops to induce ringing. The sequences were rated qualitatively by two radiologists in the categories artifacts, image sharpness, lesion conspicuity, capsule delineation and overall image quality as previously described9. Image sharpness was quantitatively assessed by measuring the apparent signal-to-noise (aSNR: SIperipheral zone/SDmuscle) and contrast-to-noise ratio (aCNR: (SIperipheral zone-SImuscle)/SDmuscle), as well as the edge rise distance across the dorsal prostate capsule. The diagnostic confidence in PI-RADS scores for each sequence was rated separately on Likert-items from 1 (low) to 5 (high). Results are given as mean and standard deviation (continuous data) or median and interquartile range (ordinal data) as appropriate. Statistically analysis was conducted using Friedman test and one-way ANOVA. A P-value of <0.05 was considered as statistically significant.Results
56 male patients were included in this ongoing study. Mean age was 67±8 years (range: 51–88 years) with a mean prostate-specific-antigen of 11.3±19.9 ng/ml. T2AI was rated significantly better in comparison to the conventionally acquired sequences in the categories image sharpness, lesion conspicuity, capsule delineation and overall image quality (Tab. 1 and 2, Fig. 1 and 2). However, a difference in appearance of artifacts was not observed. Overall agreement between both raters was high with an intercorrelation coefficient of 0.862. Quantitative analysis (Fig. 3) showed a slightly higher aSNR for T2AI (43.0±8.1) compared to T2HR (42.0±8.3, P=0.009) and T2PR (24.0±4.7, P<0.001), but not compared to T2LR (43.1±8.1, P=0.54). The aCNR of T2AI (38.2±7.9) was higher than all conventional sequences (T2LR: 37.8±7.9, P=0.017; T2HR: 35.9±8.3, P<0.001; T2PR: 19.7±4.8, P<0.001). The edge rise distance as a quantifiable measurement of image sharpness was significantly lower with 0.824±0.379 mm for T2AI (T2LR: 1,117±0.412 mm, P<0.001; T2HR: 1.445±0.789 mm, P<0.001; T2PR: 1.176±0.797 mm, P=0.001). Acquisition duration of T2LR, from which T2AI is reconstructed, is reduced to 163±21s (T2HR: 258±34s, P<0.001; T2PR: 230±30s, P<0.001). Diagnostic confidence in PI-RADS scores was higher for T2AI with a median of 4 [3-4] compared to T2LR (3 [3-4], P=0.004), T2HR (2.5 [2-3], P<0.001), and T2PR (2 [2-3], P<0.001)(Fig. 2).Discussion
Compared to the standard cartesian and propeller sequences, the new AI-reconstructed sequences show a significant improvement in nearly all aspects of image quality. This is both confirmed by a thorough qualitative and quantitative assessment. Furthermore, as the reconstruction of the AI-sequences starts from rapidly acquired low-resolution images, it effectively reduces the scan time between 37% (compared to T2HR) and 29% (compared to T2PR). However, the most important finding is the potential for an increased diagnostic performance, as a clear delineation of prostate lesions leads to a higher confidence in PI-RADS scores and therefore directly influences further diagnostics, most notably the decision for prostate biopsy. It must be noted in this context that we only investigated T2w-sequences and not diffusion-weighted sequences, which are equally important for the calculation of the PI-RADS score.Conclusion
The evaluated AI-reconstructed T2w-sequences have high potential for significantly improving diagnostic workup of patients with prostate cancer by enhancing image quality, reducing scan time and resulting in a higher confidence of PI-RADS scores. In addition, this reconstruction technique is not limited to mpMRI and can be potentially applied to all MRI-sequences after initial image acquisition. However, a thorough investigation of different applications needs to be done to ensure high quality standards.Acknowledgements
The present work was supported by Philips Healthcare.References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021 May; 71(3):209-249. DOI: 10.3322/caac.21660. Epub 2021 Feb 4. PMID: 33538338.
- Martin E, Fredrik J, Andrea D, et al. MRI-Targeted or Standard Biopsy in Prostate Cancer Screening. N Engl J Med. 2021; 385:908-920, DOI: 10.1056/NEJMoa2100852
- Hosny A, Parmar C, Quackenbush J, et al. Artificial intelligence in radiology. Nat Rev Cancer. 2018 Aug;18(8):500-510. doi: 10.1038/s41568-018-0016-5. PMID: 29777175; PMCID: PMC6268174.
- ACR, ESUR and AdMeTech Foundation. Prostate Imaging Reporting & Data System (PI-RADS). 2019.
- Pezzotti N, de Weerdt E, Yousefi S, et al. Adaptive-CS-Net: FastMRI with Adaptive Intelligence. arxiv. 2019;(NeurIPS).
- Hans Peeters, Hayley Chung, Giuseppe Valvano, et al. Philips SmartSpeed. No compromise Image quality and speed at your fingertips.
- Super-Resolution Using Deep Convolutional Networks. Chao Dong, Chen Change Loy, Kaiming He, Xiaoou Tang. 2014 arXiv:1501.00092.
- Review of the deep learning methods for medical images super resolution problems. Y. Li, B. Sixou, F.Peyrin. IRBM, Volume 42, Issue 2, April 2021, Pages 120-133.
- Bischoff LM, Katemann C,
Isaak A, et al. T2 Turbo Spin Echo With
Compressed Sensing and Propeller Acquisition (Sampling k-Space by
Utilizing Rotating Blades) for Fast and Motion Robust Prostate MRI:
Comparison With Conventional Acquisition Invest Radiol. 2022;10.1097/RLI.0000000000000923.
doi:10.1097/RLI.0000000000000923
Figures
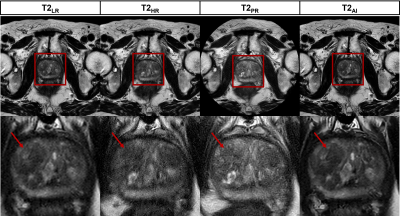
Figure 1:
Representative images for low resolution (T2LR) and high resolution
cartesian (T2HR), non-cartesian (T2PR) and
AI-reconstructed (T2AI) sequences. The red arrow marks a PI-RADS 3
lesion in the anterior transitional zone that is most sharply delineated in the
AI-recontruction.
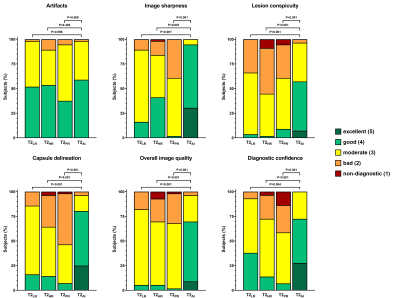
Figure 2: Bar graphs show the distribution of ratings
for each category of one rater.
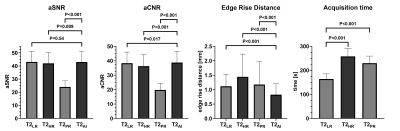
Figure 3: Quantitative analysis shows increased apparent
signal-to-noise (aSNR) and contrast-to-noise ratio (aCNR) for T2AI,
as well as a decreased edge rise distance and an reduced acquisition time.
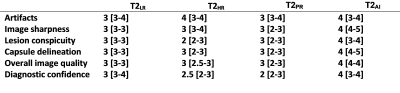
Table 1:
Quality ratings of the categories artifacts, image sharpness, lesion
conspicuity, capsule delineation, overall image quality and diagnostic
confidence on Likert items from 1 (low) to 5 (high). Median and interquartile
range are displayed.
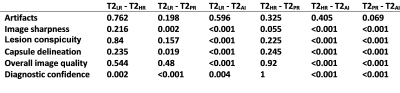
Table 2:
P-values of statistical analysis for qualitative ratings using the Friedman
test
DOI: https://doi.org/10.58530/2023/1648