1644
Automatic Segmentation of Liver Metastases Based on a Deep Learning: Assessment of Tumor Treatment Response According to the RECIST 1.1 Criteria1peking university first hospital, Beijing, China
Synopsis
Keywords: Liver, Cancer
This retrospective study aims to develop an automated algorithm for segmentation of liver metastases based on a deep learning method and assess its efficacy for treatment response assessment according to the RECIST 1.1 criteria. One hundred and sixteen treated patients with clinically confirmed liver metastases were enrolled. A 3D U-Net algorithm was trained for automated liver metastases segmentation and treatment response assessment. The results demonstrated that the automated liver metastases segmentation was capable of evaluating treatment response, with comparable results to the junior radiologist and superior to that of the fellow radiologist.Introduction
Introduction: Evaluation of treated tumors according to Response Evaluation Criteria in Solid Tumors (RECIST) criteria is an important but time-consuming task in medical imaging. Deep learning methods are expected to automate the evaluation process and improve the efficiency of imaging interpretation.Objective
Objective: To develop an automated algorithm for segmentation of liver metastases based on a deep learning method and assess its efficacy for treatment response assessment according to the RECIST 1.1 criteria.Methods
Methods: One hundred and sixteen treated patients with clinically confirmed liver metastases were enrolled. All patients had baseline and post-treatment MR images. They were divided into an initial (n = 86) and validation cohort (n = 30) according to the examined time. The metastatic foci on DWI images were annotated by two researchers in consensus. Then the treatment responses were assessed by the two researchers according to RECIST 1.1 criteria. A 3D U-Net algorithm was trained for automated liver metastases segmentation using the initial cohort. Based on the segmentation of liver metastases, the treatment response was assessed automatically with a rule-based program according to the RECIST 1.1 criteria. The segmentation performance was evaluated using the Dice similarity coefficient (DSC), volumetric similarity (VS), and Hausdorff distance (HD). The area under the curve (AUC) and Kappa statistics were used to assess the accuracy and consistency of the treatment response assessment by the deep learning model and compared with two radiologists [attending radiologist (R1) and fellow radiologist (R2)] in the validation cohort.Results
Results: In the validation cohort, the mean DSC, VS, and HD were 0.85 ± 0.08, 0.89 ± 0.09, and 25.53 ± 12.11 mm for the liver metastases segmentation. The accuracies of R1, R2 and automated segmentation-based assessment were 0.77, 0.65, and 0.74, respectively, and the AUC values were 0.81, 0.73, and 0.83, respectively. The consistency of treatment response assessment based on automated segmentation and manual annotation was moderate [K value: 0.60 (0.34-0.84)].Discussion
Discussion: Patients with liver metastases commonly require a long and regular imaging follow-up to evaluate the impact of the therapies. Size measurement is an important part of MR examination interpretation and a component of therapeutic decisions. The reproducibility of size measurements and their optimization is thus an important issue. However, some of the previous studies reported that RECIST might significantly underestimate or overestimate the disease progression due to the poor inter- and intra-observer agreement in tumor quantity[1].To overcome the limitations of manual tumor response assessment method, we proposed a deep learning-based method to segment the metastatic lesion and calculate the tumor size. Through reliable measurements of hepatic metastases, deep learning-based quantification might improve RECIST criteria performance. The 3D U-Net algorithm has been widely employed for accurate and efficient organ and tumor segmentation such as brain metastases [2-3] and liver metastases [4]. In this study, we obtained satisfied liver metastases segmentation which seems higher than the semi-automatic liver metastases segmentation on CT images performed by Eugene Vorontsov (DSC values of 0.14, 0.53, and 0.68 for the metastatic lesion smaller than 10 mm, 10-20 mm, and larger than 20 mm)[5].Conclusion
Conclusion: The deep learning-based liver metastases segmentation was capable of evaluating treatment response according to RECIST 1.1 criteria, with comparable results to the junior radiologist and superior to that of the fellow radiologist.Acknowledgements
The authors gratefully acknowledge the technical support of Yaofeng Zhang and Jialun Li from Beijing Smart Tree Medical Technology Co. Ltd.References
1. Bonekamp D, Bonekamp S, Halappa VG, Geschwind JF, Eng J, Corona-Villalobos CP, et al. Interobserver agreement of semi-automated and manual measurements of functional MRI metrics of treatment response in hepatocellular carcinoma. Eur J Radiol. 2014;83(3):487-96.
2. Bousabarah K, Ruge M, Brand JS, Hoevels M, Rueß D, Borggrefe J, et al. Deep convolutional neural networks for automated segmentation of brain metastases trained on clinical data. Radiation oncology (London, England). 2020;15(1):87.
3. Park YW, Jun Y, Lee Y, Han K, An C, Ahn SS, et al. Robust performance of deep learning for automatic detection and segmentation of brain metastases using three-dimensional black-blood and three-dimensional gradient echo imaging. Eur Radiol. 2021.
4. Goehler A, Harry Hsu TM, Lacson R, Gujrathi I, Hashemi R, Chlebus G, et al. Three-Dimensional Neural Network to Automatically Assess Liver Tumor Burden Change on Consecutive Liver MRIs. Journal of the American College of Radiology : JACR. 2020;17(11):1475-84.
5. Vorontsov E, Cerny M, Régnier P, Di Jorio L, Pal CJ, Lapointe R, et al. Deep Learning for Automated Segmentation of Liver Lesions at CT in Patients with Colorectal Cancer Liver Metastases. Radiol Artif Intell. 2019;1(2):180014.
Figures
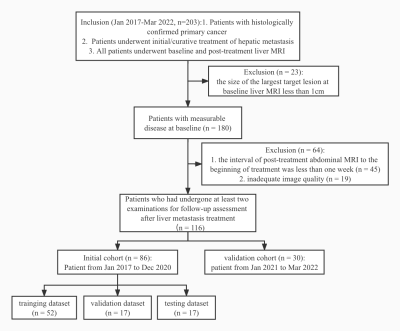
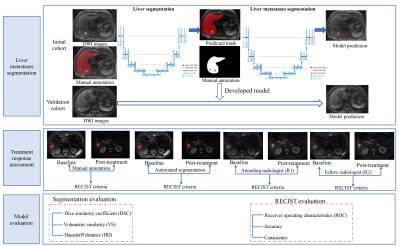
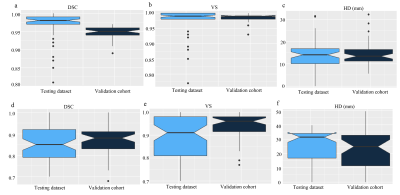
Figure 3. Notched box plots of the segmentation results in the testing dataset and validation cohort
a-c: the DSC, VS, and HD of liver segmentation; d-f: the DSC, VS and HD of liver metastases segmentation. DSC: Dice similarity coefficient; HD: Hausdorff distance; VS: Volumetric similarity.
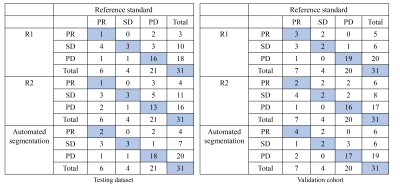
Figure 4. The confusion matrix of the response assessment results with respect to reference standard
R1: attending radiologist; R2: fellow radiologist.
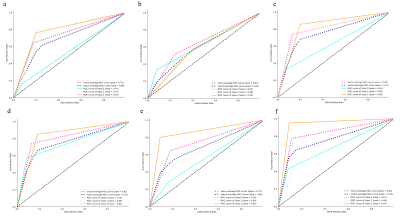
Figure 5. Receiver operating characteristic curves (ROC) for the therapy response assessment
a: attending radiologist (R1) in the testing dataset; b: fellow radiologist (R2) in the testing dataset; c: automated segmentation-based assessment in the testing dataset; d: R1 in the validation cohort; e: R2 in the validation cohort; f: automated segmentation-based assessment in the validation cohort.