1632
CMAPSS-NET: Deep-Learning Based Reconstruction of 3D Magnetization-Prepared GRE with Complementary Phase-Cycling Acquisition1Philips Healthcare, New York, NY, United States, 2Memorial Sloan Kettering Cancer Center, New York, NY, United States, 3Philips Healthcare, Boston, MA, United States, 4Albert Einstein College of Medicine and Montefiore Medical Center, Bronx, NY, United States
Synopsis
Keywords: Quantitative Imaging, Image Reconstruction
Deep learning enables efficient reconstruction of 3D Magnetization-Prepared GRE with Complementary Phase-Cycling to remove both undersampling artifacts as well as T1 contamination associated with this type of acquisition only requiring half the acquisition time compared to current standard technique.
INTRODUCTION
Magnetization-prepared (MP) GRE sequences have been utilized to improve the imaging speed in 3D quantitative T1ρ parameter mapping. However, this type of acquisition suffers from growing T1 recovery contamination and loss of MP contrast along the GRE readout train. 3D magnetization-prepared angle-modulated partitioned k-space spoiled gradient echo snapshots (MAPSS) sequence overcomes these limitations by repeating magnetization preparation with opposite (positive and negative) phase cycling (PC) during acquisition and performing subtraction after reconstruction [1]. However, this approach doubles scan time and is prone to motion artifacts. It has been recently shown that complementary PC within a single 3D data acquisition can also lead to high-resolution 3D images using compressed sensing reconstruction [2]. In this work, we present a deep learning (DL) reconstruction framework to learn the mapping between undersampled PC datasets (DL input) and MAPSS sequence images (DL reference for training) to reconstruct contamination-free images with only half the required acquisition time compared to current standard technique.METHODS
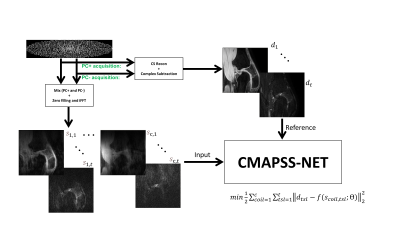
3D MAPSS sequences was acquired on healthy volunteers (n=10) on 3.0T Philips Elition X and Ingenia scanners with 1Tx/16Rx knee coil. The 3D T1ρ MAPSS scan was performed in the sagittal plane with the following scan parameters: FOV = 140×140×140 mm3, acquisition voxel size = 0.45×0.72×4 mm3 reconstructed to 512×512×30 matrix size, TR/TE = 7.3/3.7 ms, and readout bandwidth = 64.4 kHz, compressed sense acceleration factor=4. The frequency-encoding direction was the foot-head direction. A spectral selective RF pulse was performed before the T1ρ preparation module for fat suppression. Each acquisition with positive and negative PC [TSL=±0, ±10, ±20, ±30, ±40, ±50, ±60, and ±70 ms] took 35.7 s, leading to a total scan duration of 9 min 32 s for 16 acquisitions [2].DL Reference Generation: The vendor-specific wavelet-based compressed sense reconstruction algorithm on the scanner was used to generate the complex 3D images per TSL (spin-lock time). Next, each ±PC 3D dataset pair was subtracted to generate a total of 3D complex datasets free of T1 relaxation contaminations, leading to 8 datasets with different TSLs from 16 3D acquisitions as reference (Figure 1).
Input: Complex k-space data from ±PC acquisitions were retrospectively combined by selecting half of +PC and half of -PC k space readout profiles in an interleaved fashion in the ky-kz plane. One combined, each k-space was zero filled before transforming into image space using IFFT operation (Figure 1).
Deep Learning: Fully convolutional neural network with contracting and expanding paths was designed. The contracting path included 6 blocks each consisting of convolution (6×6), activation function (ReLu), batch normalization and max pooling (6×6) [3]. The architecture of the expanding path was similar except max pooling is replaced with upsampling and concatenation of corresponding feature maps with the contracting path. Dynamics (TSL=1:t) were added as additional channels. Each slice data were normalized along the coil and TSL directions. The following cost function was minimized during training
$$min\frac{1}{2}\sum_{coil=1}^c \sum_{tsl=1}^t ||d_{tsl}-f(s_{coil,tsl};\ominus)||_2^2$$
where $$$s_{coil,tsl}$$$ is the undersampled multicoil (c coils) multi dynamic (t TSLs) images, $$$d_{tsl}$$$ is reconstructed coil combined images and $$$\ominus$$$ corresponds to network weights.
Network parameters included Adam optimizer, learning rate=10-3, batch size=4, epochs=200. Data (X=256, Y=256, Z=332, Dynamic=8, Coil=10) was split into 90% for training/validation and 10% for testing. Training was performed on NVIDIA Tesla V100-SXM2-32GB with ~3 minutes per epoch.
To compare the reference with the network output, peak signal-to-noise ratio (PSNR) and structural Similarity Index (SSIM) are reported. In addition, qualitative impression, and ROI analysis along TSL are provided.
RESULTS
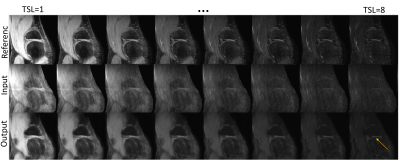
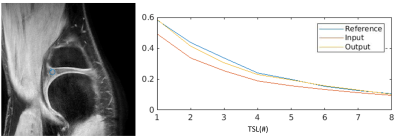
Figure 2 shows a comparison of images between reference (top), network input (middle) and network output (bottom) across TSLs. The network output images had both improved image quality and sharper contrast compared with the network input images obtained from IFFT. However, network output images were generally blurrier compared to the reference images and some of the details including vessel structures in the fat region are missing. Despite that, undersampling artifacts seen in input images are mostly removed in the DL output. In addition, cartilage structures that are barely visible in later TSLs (due to low SNR) in the input images are recovered in DL output. Corresponding PSNR and SSIM values for Figure 2 are shown in Table 1.In Figure 3, ROI analysis along TSL shows improved agreement between the reference and DL output compared to that of input where the starting point is much lower, and signal decay much slower, suggesting a different T1ρ.
DISCUSSION and CONCLUSION
This work shows feasibility of using deep learning to reconstruct undersampled, T1 contamination-free images using only half the data typically required in standard approach. This allows 50% of scan duration reduction and potentially facilitates the application of quantitative T1ρ mapping in routine clinical practice. Results can further be improved by including more training cases. In addition, motion was observed in a few subjects between datasets at different TSLs. Therefore, pre-training motion correction will further improve the network performance. While the network was trained with all TSLs per slice, to implicitly seek correlation along dynamic/time, it is also possible to train the network TSL by TSL to minimize motion artifact contribution during training.Acknowledgements
No acknowledgement found.References
[1] Li X, Han ET, Busse RF, Majumdar S. In vivo T1ρ mapping in cartilage using 3D magnetization-prepared angle-modulated partitioned k-space spoiled gradient echo snapshots (3D MAPSS). Magn Reson Med 2008; 59(2): 298-307.
[2] Peng Q, Zhao Y, Wu C, “Fast High-Resolution 3D Magnetization-Prepared GRE with Complementary Phase-Cycling Acquisitions”, AAPM Annual Meeting, Washington DC, 2022.
[3] Jafari, R, Spincemaille, P, Zhang, J, et al. Deep neural network for water/fat separation: Supervised training, unsupervised training, and no training. Magn Reson Med. 2020; 85: 2263– 2277.
Figures