1628
Integrating Adaptive Generative Network and Subspace Models for Accelerated MR Parameter Mapping1Beckman Institute for Advanced Science and Technology, Urbana, IL, United States, 2Department of Electrical and Computer Engineering, University of Illinois Urbana-Champaign, Urbana, IL, United States, 3Department of Radiology, Mayo Clinic, Rochester, MN, United States, 4Department of Bioengineering, University of illinois Urbana-Champaign, Urbana, IL, United States
Synopsis
Keywords: Quantitative Imaging, Machine Learning/Artificial Intelligence
We proposed a new method that integrates an adapted generative network image prior and subspace modeling for accelerated MR parameter mapping. Specifically, a formulation is introduced to synergize a subspace constraint, a subject-specific generative model-based image representation and joint sparsity regularization. A pretraining using public database plus subjective specific network adaptation strategy is used to construct an accurate representation of the unknown contrast-weighted images. An efficient alternating minimization algorithm is used to solve the resulting optimization problem. The improved reconstruction performance achieved by the proposed method over subspace and sparsity constraints was demonstrated in a T2 mapping experiment.Introduction
Quantitative MR parameter mapping (qMR) is playing an ever-important role in tissue characterization and pathological analysis1-2. One major challenge for clinical application of qMR is the prolonged acquisition due to the need to acquire multiple contrast-weighted images for parameter estimation. Image reconstruction from sparsely sampled data has demonstrated excellent potential in accelerating qMR. For example, constrained reconstructions using sparse representations3-4, low-rank matrix/tensor models5-8, and their combinations have been well recognized. Recently, deep learning (DL)-based methods have provided new ways to formulate qMR problems and to introduce data adaptive priors. The majority of existing DL methods rely on supervised training using a large number of fully-sampled datasets9-10. To address the data scarcity issue, self-supervised learning strategies have also shown promise11-12. In this work, we proposed a new method that integrates a state-of-the-art generative image model and subspace model for qMR reconstruction. We introduced a pretraining plus subject-specific adaptation strategy for learning an accurate generative adversarial network (GAN)-based image representation and used it as an adaptive spatial constraint in a subspace-based reconstruction. Our method does not require end-to-end supervised training and allows flexible integration of other constraints, e.g., joint-sparsity, to further improve the reconstruction. We demonstrated the effectiveness of our method using T2 mapping experiments.Theory and Methods
Generative prior constrained subspace reconstructionWe formulate the qMR reconstruction problem as follows:
$$\hat{\mathbf{U}},\hat{\mathbf{w}}_{\text {rec}}=\text{arg}\underset{\mathbf{U},\mathbf{w}}{\operatorname{min}}\sum_{c=1}^{N_c}\left\|\mathbf{d}_{\mathbf{c}}-\boldsymbol{\Omega}\left(\mathbf{F S}_{\mathbf{c}}\mathbf{U}\hat{\mathbf{V}}\right)\right\|_2^2+\lambda_1\left\|\mathbf{U} \hat{\mathbf{V}}-\boldsymbol{G}_{\boldsymbol{\hat{\theta}}}(\mathbf{w})\right\|_F^2+\lambda_2 R(\mathbf{U}\hat{\mathbf{V}}),\text{(1)}$$
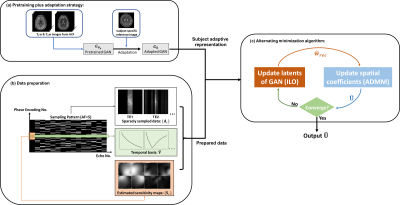
where $$$\boldsymbol{\Omega},\mathbf{F},\mathbf{S}_{\boldsymbol{c}},\mathbf{d}_{\mathbf{c}},\hat{\mathbf{V}}$$$ denote the $$$(\mathrm{k},\mathrm{p})$$$-space sparse sampling mask ( $$$\mathrm{p}$$$ denoting the parameter dimension, e.g., TE in T2 mapping), Fourier operator, coil-dependent sensitivity maps, measured multichannel data, and a predetermined temporal basis, respectively. The first term in Eq. (1) is the well-established subspace/low-rank model, and $$$R(.)$$$ can be a hand-crafted regularization term, e.g., promoting joint sparsity4,5. The second term introduces a GAN-based image-manifold constraint. One key assumption is that for a well-trained and adapted GAN-based image model with fixed parameter $$$\hat{\boldsymbol{\theta}}$$$, the contrast variations in the image sequence can be accurately accounted for by updating only the low-dimensional latent space variable $$$\mathbf{w}$$$ (referred to as the latents). Here, we propose a pretraining plus subject-specific adaptation strategy to construct $$$\boldsymbol{G}_{\hat{\boldsymbol{\theta}}}$$$ without needing a large scale T2 mapping dataset for training.
Adaptive generative image model
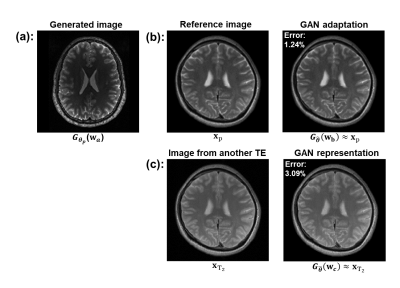
To construct a generative model that can capture geometry and contrast variations in brain MRI, we first pretrain a StyleGAN213(excellent high-resolution image generation capability), on the HCP database14 ( ~100K T1w&T2w images). Assume that a high-resolution reference image $$$\mathbf{x}_{\mathrm{p}}$$$ is available in a parameter mapping experiment (not necessarily the same contrast), we can adapt the pretrained $$$\boldsymbol{G}_{\boldsymbol{\theta}_{\mathbf{p}}}(.)\left(\boldsymbol{\theta}_{\mathbf{p}}\right.$$$ denotes the network parameters) to $$$\mathbf{x}_{\mathbf{p}}$$$ by updating both the network parameters and latents $$$\mathbf{w}$$$ as:
$$\hat{\mathbf{w}}, \hat{\boldsymbol{\theta}}=\text{arg}\underset{\mathbf{w},\boldsymbol{\theta}}{\operatorname{min}}\left\|G_{\boldsymbol{\theta}}(\mathbf{w})-\mathbf{x}_{\mathbf{p}}\right\|_2^2+\alpha\left\|\boldsymbol{\theta}-\boldsymbol{\theta}_{\mathbf{p}}\right\|_2^2.\text{(2)}$$
The second term penalizes large deviation of the adapted network from the pretrained one. This pretraining+adaptation strategy produces an effective GAN-based image prior for which updating only the low-dimensional latents can accurately account for contrast variations, as shown in Fig. 1. A strategy that uses a T1-to-T2 image translation network to generate reference image for constrained reconstruction was previously proposed15. In our method, the network serves as an image representation that is dynamically updated in a unified optimization process (Eq. (1)).
Algorithm
With the adapted $$$\boldsymbol{G}_{\hat{\boldsymbol{\theta}}}$$$ and $$$R(.)=\|\mathbf{D U \hat { V }}\|_{\mathbf{2,1}}$$$ ($$$\mathbf{D}$$$ being a finite difference operator5 ), we solve Eq. (1) using an alternating minimization algorithm. More specifically, the algorithm alternates between updating the latents $$$\mathbf{w}$$$ and spatial coefficients $$$\mathbf{U}$$$, i.e.,$$\begin{gathered}\hat{\mathbf{w}}_{rec}^{i+1}=\text{arg}\underset{\mathbf{w}}{\operatorname{min}}\left\|\hat{\mathbf{U}}^i\hat{\mathbf{V}}-G_{\hat{\boldsymbol{\theta}}}(\mathbf{w})\right\|_F^2;\text{(3)}\\\hat{\mathbf{U}}^{i+1}=\text{arg}\underset{\mathrm{U}}{\operatorname{min}} \sum_{c=1}^{N_c}\left\|\mathrm{~d}_{\mathbf{c}}-\Omega\left(\mathbf{F S}_{\mathrm{c}}\mathbf{U}\hat{\mathbf{V}}\right)\right\|_2^2+\lambda_1\left\|\mathbf{U} \hat{\mathbf{V}}-G_{\hat{\theta}}\left(\hat{\mathbf{w}}_{\text{rec }}^{i+1}\right)\right\|_F^2+\lambda_2\|\mathbf{X}\|_{2,1}, \text{s.t.}\mathbf{X}=\mathbf{DU\hat{\mathbf{V}}};\text{(4)}\end{gathered}$$
where $$$i$$$ is the iteration index and $$$\mathbf{X}$$$ an auxiliary variable for handling the $$$l_{2,1}$$$-norm. We used the intermediate layer optimization (ILO) algorithm16 to solve (3) and ADMM17 to solve (4). The algorithmic workflow is illustrated in Fig. 2.
Results
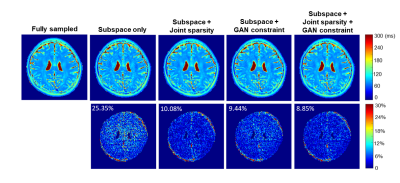
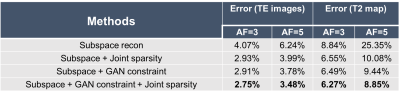
We evaluated the proposed method using in vivo T2 mapping data (with local IRB approval). Data were acquired using a multi-spin-echo sequence on a 3T scanner using a 12-channel head coil: 16 echoes, $$$\mathrm{TE}_1=8.8 \mathrm{~ms}$$$ and echo spacing $$$\Delta \mathrm{TE}=8.8 \mathrm{~ms}$$$. The proposed method was benchmarked against a state-of-the-art joint subspace and sparsity constrained reconstruction using retrospectively under-sampled data at different acceleration factors (AFs). The central 8 k-space lines were acquired at all TEs for subspace determination5,6, and the central k-space was fully sampled at 1st TE (coverage depending on AFs, e.g., 32 $$$k_y$$$'s for AF = 4) for coil sensitivity estimation by ESPIRiT18.For T2 mapping, we used a sum-of-squares (SoS) combination of images across all TEs from an initial subspace reconstruction as $$$\mathbf{x}_{\mathrm{p}}$$$ for adapting the pretrained StyleGAN2 (Eq. (2)); note that the SoS image exhibits negligible aliasing even at high AFs. While a high-resolution T1w image can also be used, the strategy here may eliminate additional acquisitions. Fig. 3 compares the reconstruction from different methods at AF=5, demonstrating the superior performance of the proposed method, especially when combining joint sparsity constraint. More quantitative comparisons are shown in Table 1. The proposed method consistently achieved lower errors at different AFs.
Conclusion
We proposed a novel reconstruction method for qMR that integrates subspace modeling and a subject-adaptive GAN-based image representation. We validated the accuracy of our pretrained and adapted StyleGAN2 in capturing contrast variations, and demonstrated improved reconstruction by the proposed method for accelerated T2 mapping.Acknowledgements
This work was supported in part by NSF-CBET-1944249 and NIH-NIBIB-1R21EB029076AReferences
1.Ma, Dan, et al. Magnetic resonance fingerprinting. Nature 2013; 495:187-192.
2.Alexander, Andrew L., et al. Characterization of cerebral white matter properties using quantitative magnetic resonance imaging stains. Brain connectivity 2011;1: 423-446.
3.Velikina, Julia V., Andrew L. Alexander, and Alexey Samsonov. "Accelerating MR parameter mapping using sparsity‐promoting regularization in parametric dimension." MRM 2013; 70: 1263-1273.
4.Doneva, Mariya, et al. "Compressed sensing reconstruction for magnetic resonance parameter mapping." MRM 2010; 64: 1114-1120.
5.Zhao, Bo, et al. "Accelerated MR parameter mapping with low‐rank and sparsity constraints." MRM 2015; 74: 489-498.
6.Peng, Xi, et al. "Accelerated exponential parameterization of T2 relaxation with model‐driven low rank and sparsity priors (MORASA)." MRM 2016; 76: 1865-1878.
7.He, Jingfei, et al. "Accelerated high-dimensional MR imaging with sparse sampling using low-rank tensors." IEEE transactions on medical imaging 2016; 35: 2119-2129.
8.Christodoulou, Anthony G., et al. "Magnetic resonance multitasking for motion-resolved quantitative cardiovascular imaging." Nature biomedical engineering 2018; 2: 215-226.
9.Liu, Fang, Li Feng, and Richard Kijowski. "MANTIS: Model‐Augmented Neural neTwork with Incoherent k‐space Sampling for efficient MR parameter mapping." MRM 2019; 82: 174-188.
10.Hamilton, Jesse I., et al. "Deep learning reconstruction for cardiac magnetic resonance fingerprinting T1 and T2 mapping." MRM 2021; 85: 2127-2135.
11.Cheng, Jing, et al. "Deep MR Parametric Mapping with Unsupervised Multi-Tasking Framework." Investigative Magnetic Resonance Imaging 2021; 25: 300-312.
12.Liu, Fang, et al. "Magnetic resonance parameter mapping using model‐guided self‐supervised deep learning." MRM 2021; 85: 3211-3226.
13. Karras, Tero, et al. "Analyzing and improving the image quality of stylegan." Proceedings of the IEEE/CVF conference on computer vision and pattern recognition. 2020.
14.Van Essen, David C., et al. "The WU-Minn human connectome project: an overview." Neuroimage 2013; 80: 62-79.
15.Meng, Ziyu, et al. "Accelerating T2 mapping of the brain by integrating deep learning priors with low‐rank and sparse modeling." MRM 2021; 85: 1455-1467. 16.Daras, Giannis, et al. "Intermediate Layer Optimization for Inverse Problems using Deep Generative Models." ICML. PMLR, 2021.
17.Boyd, Stephen, et al. "Distributed optimization and statistical learning via the alternating direction method of multipliers." Foundations and Trends® in Machine learning 2011; 3: 1-122.
18.Uecker, Martin, et al. "ESPIRiT—an eigenvalue approach to autocalibrating parallel MRI: where SENSE meets GRAPPA." MRM 2014; 71: 990-1001.
Figures