1613
Simulating the efficiency of Variable Flip Angle (VFA) multi-parametric mapping of T1, PD, and T2* at 7T suggests longer TRs may be optimal1Biomedical Engineering Department, School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 2London Collaborative Ultra high field System (LoCUS), London, United Kingdom, 3MR Research Collaborations, Siemens Healthcare Limited, Frimley, United Kingdom, 4London Collaborative Ultra high field System (LoCUS), Lodnon, United Kingdom, 5Centre for the Developing Brain, School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom
Synopsis
Keywords: Quantitative Imaging, Neuro, MPM, sequence optimization
Quantitative MRI at 7T may have important clinical applications owing to the potential for enhanced resolution and uniform tissue characterisation. Current protocols, often for pragmatic reasons, use short TRs(~20ms) and a handful of echo times but this might not be optimal for T2* estimation. We simulated the optimal TR and echo number for dual flip angle mapping of T1, PD, and T2* for realistic 7T tissue and sequence parameters. The optimal efficiency was found to be at 65ms with 20 echoes. The simulations suggested that longer TRs of 40-60ms would be optimal. Preliminary in-vivo data only partially supported these findingsIntroduction
Quantitative MRI at 7T may have important clinical applications owing to the potential for enhancedresolution and uniform tissue characterisation. Variable flip angle (VFA)[1] a spoiled gradient echo (SPGR) approaches such as Multi-Parametric Mapping (MPM)[2] are robust at 7T however, protocol optimisation to achieve precise parameter mapping across the whole brain with high resolution remains challenging due to large variability in transmit field (B1+). In this abstract, for a dual flip angle multi-echoSPGR acquisition, we aimed to optimise T1,T2* and PD map precision for protocol parameters: flip angle, TR and number of echoes, accounting for realistic B1 variability using the Cramer-Rao Lower Bound (CRLB) approach.
Methods
Simulation: A multi-echo SPGR sequence was simulated using the Ernst equation approximation[3] as a function of tissue parameters T1,T2* and PD. Scanner protocol parameters: flip-angle (α), TR,TE considered in the simulation were selected within sequence-permitted ranges. To calculate the achievable range of number of echoes and corresponding TE times for a given TR within hardware constraints, the receiver bandwidth was varied from 1 to 1780Hz/Px. Corresponding readout gradients and ramp times were calculated for FOV=256mm at 1mm isotropic keeping within scanner limits (80mT/m & 195T/m/s). Maximum inter-echo spacing (ΔTE) and minimum readout bandwidth (BWreadout)was calculated aiming to minimise dead time. The ΔTE was constant for all echoes at a given TR.Coefficient of variation (CoV) was estimated using the CRLB framework[4], for T1 grid 1220ms,2130ms, T2* grid 14ms,53ms PD grid 0.69 and 0.82(a.u.) corresponding to white matter and gray matter, respectively. Gaussian noise with a standard deviation of σtheor=0.02 was assumed (SNR~35) based on a reference PDw image with BWreadout=230Hz/Px. The σtheor SNR values were then scaled by according to the relative BWreadout to account for SNR effects. The worst case (across tissues) CoV was minimized over a range of flip angles α1,α2∈{1°,60°}, TR ∈{15ms,100ms} and TE∈{1.33ms,98ms} for T1, PD and T2* individually. B1+ variability was incorporated by summing the worst-case CoV calculated by setting κ∈ {0.67,0.78,0.89,1.00,1.11} corresponding to the range obtained in the brain from the experimental B1+ maps. The efficiency metric in T1, PD and T2* estimation was obtained from CoVT1,CoVPD and CoVT2*[5]. The combined efficiency of estimating T1, PD and T2* was obtained by setting the cost function equal to the sum of individual CoV.
Preliminary Experimental Validation: In-vivo data was acquired in 2 adult HV subjects. B1+ homogeneity was mapped using the actual flip angle imaging method with αnom=60°[7]. Data was obtained at TR =20,40 and 60ms.(Table 1).All the image volumes were coregistered and resliced. To eliminate potential distortion effects owing to the relatively low bandwidth, only even echoes were used in the analysis of relative CoV with TR. T1,PD and T2* maps were calculated using a GLM framework[6]. Subject specific WM&GM regions were determined from probabilistic tissue segmentation in spm12(threshold p<0.9) from a reference MP2RAGE.
Results
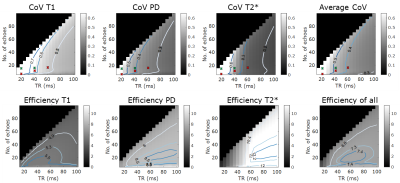
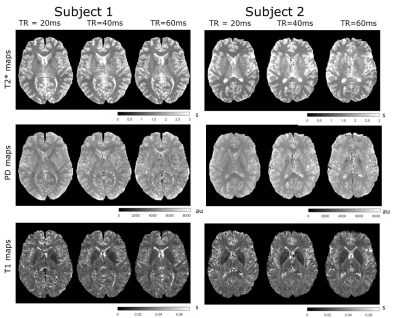
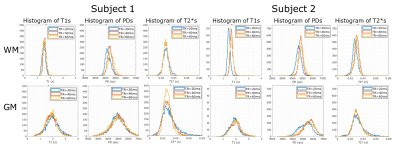
Simulations results are shown in Fig. 1. The top row shows the coefficients of variation of the T1, PD and T2* estimate respectively obtained from the CRLB framework as a function of TR and number of echoes. Bottom row shows the corresponding efficiencies of the parameters. The contours are labelled to depict 90,85,60,50 percentiles in each heatmap. The optimal flip angles for combined efficiency were found to be 6°&19° at TR=20ms, 10°&42° at TR=40ms and 11°&52° at TR=60ms. The most efficient TR for T1 estimation was calculated to be 37ms with 14 echo readoutFig. 2 depicts the experimental results for both subjects. The T1, PD and T2* maps obtained for subject 1 and subject 2 for the different TRs are shown. Representative axial images show notable changes in contrast in the T2* maps as TR and echo readouts increase. Fig. 3 shows the histogram distributions in the WM and GM ROIs from both subjects for each tissue parameter. Overall the CoV for T2* seemed to decrease as TR increased in both subjects (narrower histograms)
Discussion
Simulations showed that increasing TR and echo number should be beneficial for overall efficiency of estimating T1,PD and T2*, which is mainly driven by the increase in efficiency of T2*. Simulations include realistic gradient levels but exclude effects due to RF spoiling which may affect in-vivo data.Simulations also exclude effects due to eddy currents that maybe more problematic at higher echo numbers requiring larger gradient amplitudes. Invivodata demonstrated an effect of TR on the fitting of data from vessels which may be caused due to differential flow sensitivity. T2* experimental data quality is
hard to assess as WM and GM contain significant biological variability although some improvement in PD variability was also suggested by the histograms.These preliminary results suggested increasing
echoes and a modest TR increase might be helpful with visual improvements in the map. These findings require
replication in a larger cohort and the increased scan duration due to increased TRs would require greater image acquisition acceleration or motion compensation.Owing to the contrast changes seen with TR fitting approaches may also need further optimisation,particularly for regions with shorter T2* values.
Acknowledgements
EPSRC CDT PhDstudentship (JM). 'Wellcome/EPSRC Centre for Medical Engineering [WT203148/Z/16/Z] and by the National Institute for Health Research
(NIHR) Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation
Trust and King’s College London and/or the NIHR Clinical Research Facility. The
views expressed are those of the author(s) and not necessarily those of the
NHS, the NIHR or the Department of Health and Social Care. This research was
supported by GOSHCC Sparks Grant V4419 (SD/DC).
References
[1] Haase A, Frahm J, Matthaei KD., FLASH imaging: rapid NMR imaging using low flip angle pulses, J Magn Reson 1986: 67: 258-266
[2] Weiskopf N, Suckling J, Williams G, Correia MM, Inkster B, Tait R, Ooi C, Bullmore ET, Lutti A. Quantitative multi-parameter mapping of R1, PD(*), MT, and R2(*) at 3T: a multi-center validation. Front Neurosci. 2013 Jun 10;7:95
[3] Helms, G., Dathe, H. and Dechent, P. (2008), Quantitative FLASH MRI at 3T using a rational approximation of the Ernst equation. Magn. Reson. Med., 59: 667-672
[4] Teixeira, R.P.A., Malik, S.J. and Hajnal, J.V. (2018), Joint system relaxometry (JSR) and Crámer-Rao lower bound optimization of sequence parameters: A framework for enhanced precision of DESPOT T1 and T2 estimation. Magn. Reson. Med, 79: 234-245
[5] Leitão D, Teixeira RPAG, Price A, Uus A, Hajnal JV, Malik SJ. Efficiency analysis for quantitative MRI of T1 and T2 relaxometry methods. Phys Med Biol. 2021 Jul 26;66(15):15NT02. doi: 10.1088/1361-6560/ac101f. PMID: 34192676; PMCID: PMC8312556.
[6] Lorio S, Tierney TM, McDowell A, Arthurs OJ, Lutti A, Weiskopf N, Carmichael DW. Flexible proton density (PD) mapping using multi-contrast variable flip angle (VFA) data. Neuroimage. 2019 Feb 1;186:464-475
[7] Yarnykh VL. Actual flip-angle imaging in the pulsed steady state: a method for rapid three-dimensional mapping of the transmitted radiofrequency field. Magn Reson Med. 2007 Jan;57(1):192-200
Figures