1567
Habitat analysis based on diffusion kurtosis imaging to predict adult-type diffuse gliomas grade1Affiliated Hospital of Inner Mongolia Medical University, Hohhot, China, 2Inner Mongolia Medical University, Hohhot, China, 3Siemens Healthineers, Shanghai, China
Synopsis
Keywords: Tumors, Diffusion/other diffusion imaging techniques
Glioma composition is complicated, making whole-tumor investigation with traditional pathological analysis typically challenging. It is possible to demonstrate intratumor heterogeneity using habitat analysis in radiography, a technique analogous to biological environmental analysis with individualized descriptions and quantitative expression of subregions inside the region of interest. Therefore, our goal was to create an integrated model for the habitat analysis-based diffusion kurtosis imaging diagnosis of adult-type diffuse glioma grade. According to the findings, diffusion kurtosis imaging based on habitat analysis accurately predicted adult-type diffuse gliomas grade, which may help to resolve some "intermediate situations" that are challenging to diagnose clinically.Background and Purpose
The microenvironment of gliomas is generally recognized to be extremely complex, and pathological diagnostics typically do not allow for a thorough and all-encompassing examination. We tried to create a predictive model for spatial habitat analysis based on diffusion kurtosis imaging (DKI) to successfully detect adult-type diffuse gliomas grade to make it easier to create exact treatment strategies.Methods
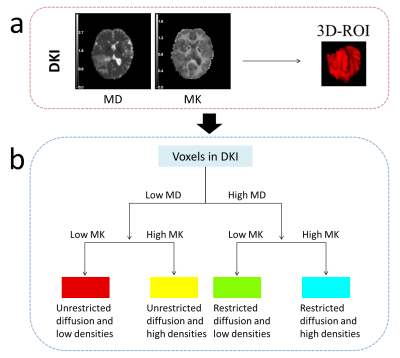
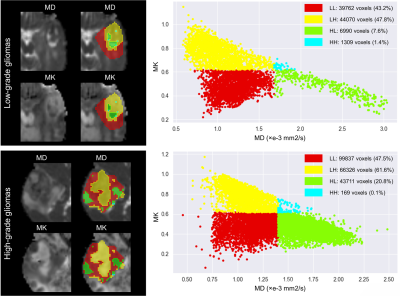
A total of 103 participants (mean age, 52 years; range, 21-77; 54 [52%] male) with adult-type diffuse gliomas were included in the secondary analysis of this prospective investigation. Participants in the study included 46 low-grade gliomas (Central Nervous System WHO Grade 2-3) and 57 high-grade gliomas (Central Nervous System WHO Grade 4). Every participant received preoperative MRI using a 3T scanner (MAGNETOM Skyra; Siemens Healthcare, Erlangen, Germany), which includes conventional MRI and diffusion spectrum magnetic resonance imaging (DSI). The DSI sequence was obtained in the axial plane using a half q-space Cartesian grid sampling procedure under the following parameters: TR/TE = 7000/107 ms, FOV = 260 mm × 260 mm, GRAPPA = 2, layer thickness = 3.0 mm, voxel size = 2.2× 2.2× 3.0mm3, 50 layers and maximum B value = 3000 s/mm2. All DSI raw data were processed using the NeuDiLab software developed in-house based on the open resource tool DIPY (Difusion Imaging in Python, https://dipy.org/) [1]. Mean diffusivity (MD) and mean kurtosis (MK) maps of DKI, as the main quantitative parameters, were used for habitat space feature extraction. For each map, we used Otsu threshold method to split the voxels into high- and low- region on the whole cohort. Then we combined these two sub-regions of each map to get the final four sub-regions, which recognized the unrestricted diffusion and low densities (LL), unrestricted diffusion and high densities (LH), restricted diffusion and low densities (HL), and restricted diffusion and high densities (HH) sub-region, respectively. Tumor entities and the edema they cause were used for analysis. Using the Mann‒Whitney U test or Student's t test, single factors with differences were found. Then, using FeAture Explorer (FAE v0.5.2, https://github.com/salan668/FAE) [2], a thorough predictive model building process was carried out with the goal of selecting the optimum model by utilizing several pipelines. Cross-validation using the leave-one-out technique was used to evaluate the stability of the model. The effectiveness of the diagnosis was assessed using ROC curves.Results
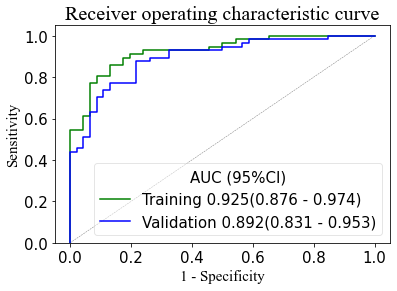
The integrated diagnostic model contained 7 single predictors, risk factors such as MD_LH, MK_LH, LL volume percentage, and LH volume percentage (odds ratios 14.978, 16.787, 2.990 and 3.912), as well as conservation factors such as MK_LL, MK_HH, and LH volume (odds ratios 43.110, 1.920, and 0.036, respectively). The combined habitat prediction model displayed a good AUC (0.925), moderate sensitivity (0.842), and specificity (0.842). Internal validation tests showed stability (AUC = 0.892).Discussion and Conclusion
This study found a relationship between the spatial parameters of the tumor habitat and the grade of adult-type diffuse gliomas. High densities and diffusion restriction typically have a favorable correlation with tumor aggressiveness. However, in actuality, a few "exceptional cases"—such as those with high diffusion restriction and low density or tumors with a histological grade between grade 2 and grade 3—frequently arise, significantly raising the level of ambiguity in clinical work. Our study's findings successfully address the "mismatch" phenomenon, enabling certain in-between situations to be classified into one of these groups. Furthermore, several earlier studies [3,4] frequently only used traditional imaging techniques and neglected to use more sophisticated diffusion models, which might offer some guidance for future multimodal analysis. Increasing the sample size or conducting the methodology correctly remain problems, of course. Finally, habitat analysis-based diffusion kurtosis imaging, a novel noninvasive analytic technique, accurately and completely predicts the grade of adult-type diffuse gliomas, and its assessment may enhance the formulation of imaging-based treatment plans.Acknowledgements
This work was supported by the Inner Mongolia Autonomous Region Science and Technology Plan Project (2019GG047). The authors gratefully acknowledge the essential contributions of the research staff of Affiliated Hospital of Inner Mongolia Medical University.References
1. Wang P, Weng L, Xie S, et al. Primary application of mean apparent propagator-MRI diffusion model in the grading of diffuse glioma. Eur J Radiol 2021;138:109622.
2. Song Y, Zhang J, Zhang YD, et al. FeAture Explorer (FAE): A tool for developing and comparing radiomics models. PLoS One. 2020;15(8):e0237587. Published 2020 Aug 17.
3. Beig N, Bera K, Prasanna P, et al. Radiogenomic-Based Survival Risk Stratification of Tumor Habitat on Gd-T1w MRI Is Associated with Biological Processes in Glioblastoma. Clin Cancer Res. 2020;26(8):1866-1876.
4. Verma R, Correa R, Hill VB, et al. Tumor Habitat-derived Radiomic Features at Pretreatment MRI That Are Prognostic for Progression-free Survival in Glioblastoma Are Associated with Key Morphologic Attributes at Histopathologic Examination: A Feasibility Study. Radiol Artif Intell. 2020;2(6):e190168. Published 2020 Nov 11.
Figures