1481
Development and validation of an MRI-based radiomics nomogram for evaluation of chronic outcome in drug-induced liver injury1Radiology, Ruijin Hospital Affiliated to Shanghai Jiao Tong University, Shanghai, China
Synopsis
Keywords: Liver, Radiomics, drug-induced liver injury
Drug-induced liver injury (DILI) occurs in a small fraction of individuals exposed to drugs, herbs or dietary supplements. DILI is challenging because of the lack of biomarkers to predict chronic outcomes. In our study, we developed a radiomics model with excellent performance for prediction of chronic outcome based on MRI and clinical data. Decision curve analysis confirmed the clinical utility of the radiomics model. This model may be used to stratify DILI patients at high risk of chronic outcome.Introduction
Drug-induced liver injury (DILI) is defined as the liver damage induced by various of drugs1, including drugs, health products, and dietary supplements, with an increasing incidence year by year. Although most liver damage of DILI patients resolve after the discontinuation of the insulting drugs, 6-39% of patients with DILI suffer persistent liver damage, which could result in cirrhosis and even liver failure. Several risk factors have been identified to be associated with the chronicity of DILI, such as older age, female sex and injury patterns. However, it remains a major challenge to accurately predict the chronic DILI because of the lacking of the predictive factor with excellent performance. Radiomics is an emerging field where quantitative radiomic features were extracted and selected using various techniques, and it could resolve clinical problems and optimize the clinical management with satisfactory potential. Radiomics have been applied in clinical practice of various liver diseases. Radiomics could classify liver hemangioma, liver metastases and hepatocellular carcinoma with satisfying diagnostic performances2. Additional, radiomics was also utilized for the prediction of NASH and the noninvasive assessment of liver fibrosis3. Based on these grounds, we speculated that radiomics could be utilized to predict the chronicity of DILI. In this study, we aimed to establish and validate a nomogram4 based on the radiomics and clinical characteristics, which could stratify DILI patients at high risk of chronic outcome.Methods
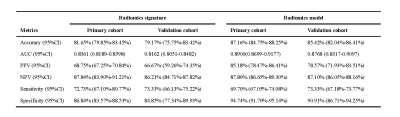
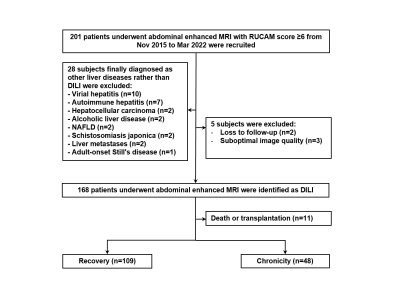
This was a retrospective single-center study, which was concordance with the Declaration of Helsinki and approved by the Ethics Committee of Shanghai Ruijin Hospital, School of Medicine, Shanghai Jiao Tong University. Written informed consent of all subjects was obtained. From Nov 2015 to Mar 2022, hospitalized DILI patients who underwent abdominal gadoxetic acid-enhanced MRI were retrospectively enrolled from the Department of Infectious Diseases, Ruijin Hospital, China. The final cohort consisted of 157 consecutive patients, with 109 cases recovered, 48 cases progressed into chronicity. MRI were obtained with either a 1.5-T or a 3.0-T MR system (Aera, Siemens; Ingenia, Philips Medical Systems; uMR 660, UIH) with the same protocol including T1WI, T2WI, DWI, and DCE with arterial, venous, and delayed phase. The liver mask was segmented automatically by using a pre-trained convolutional neural network (CNN)5. The liver mask was further amended by a radiologist with 5 years of experiences. Because fibrosis of the liver occurs during the development of DILI, only delayed phase images were used to extract radiomics features6. Patients were allocated to primary and validation cohorts randomly in a 7:3 ratio. The clinical characteristics of all patients are shown in Table 1. The radiomics features were extracted from the delayed phase image using the PyRadiomics toolbox in Python7. Clinical data were also fed into the predictive model. To avoid overfitting, t test and the least absolute shrinkage and selection operator (LASSO) regression (λ = 0.001) with leave-one-out cross-validation was applied to reduce the dimensionality of features. Finally, the support vector machine (SVM) method was used to stratify DILI patients at high risk of chronic outcome in this study. A radiomics score was calculated for each patient using an SVM model with linear kernel training based on the selected features. The Area-under-the-curve (AUC), classification accuracy, positive predictive value (PPV), negative predictive value (NPV), sensitivity and specificity were calculated as metrics to assess the quantitative discrimination performance of the radiomics signature in both the primary and validation cohorts.Results
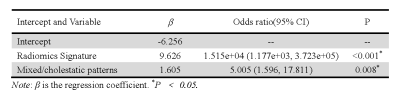
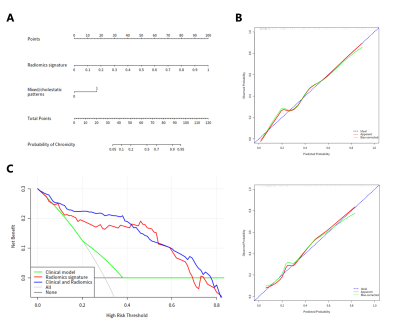
The radiomics signature comprised 28 selected features and showed good discrimination performance in both the primary and validation cohorts. The individualized radiomics model, which incorporated the radiomics signature and injury type, also showed good discrimination. There was a significant difference in radiomics scores between recovery and chronic patients in the primary cohort (P < 0.001); the same was true in the validation cohort (P < 0.001). The radiomics signature yielded an AUC of 0.8361 [95% confidence interval (CI), 0.8189–0.8598] and a classification accuracy of 81.65% (95% CI, 79.85–83.45%) in the primary cohort, and an AUC of 0.8162 (95% CI, 0.8053–0.8482) and a classification accuracy of 79.19% (95% CI, 75.75–83.42%) in the validation cohort. The radiomics signature and injury type, were identified as independent factors predicting chronic outcome. The radiomics signature and injury type were identified as independent factors predicting chronic outcome. Decision curve analysis confirmed the clinical utility of the radiomics model.Discussion and Conclusion
In this study, we propose a validated and easy-to-use radiomics model for stratifying DILI patients at high risk of chronic outcome. The easy-to-use nomogram facilitated estimation of risk of chronic outcome. The proposed radiomics model performs well and thereby provides an effective tool for clinical decision-making. Our research has several limitations. First, all the patients were from a single center. Although we categorized the patients into independent primary and validation cohorts randomly, the model may perform differently if multicenter datasets with different parameters are used. Secondly, we only use an SVM model with linear kernel for the prediction task. More recently proposed architectures such as lightBGM can be used to further improve the prediction performance. Finally, more modalities will be used for radiomics signature construction in further work.Acknowledgements
No acknowledgement found.
References
1. Kuna L, Bozic I, Kizivat T, et al. Models of drug induced liver injury (DILI)–current issues and future perspectives. Current drug metabolism. 2018;19(10);830-8.
2. Larue R T, Defraene G, De Ruysscher D, et al. Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. The British journal of radiology. 2017;90(1070);20160665.
3. Wei J, Jiang H, Gu D, et al. Radiomics in liver diseases: Current progress and future opportunities. Liver International. 2020;40(9);2050-63.
4. Zhao L, Gong J, Xi Y, et al. MRI-based radiomics nomogram may predict the response to induction chemotherapy and survival in locally advanced nasopharyngeal carcinoma. European radiology. 2020; 30(1);537-46.
5. Isensee F, Jaeger P F, Kohl S A, et al. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nature methods. 2021;18(2);203-11.
6. Wang C Y, Deng Y, Li P, et al. Prediction of biochemical nonresolution in patients with chronic drug‐induced liver injury: a large multicenter study. Hepatology. 2022;75(6);1373-85.
7. Van Griethuysen J J, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer research. 2017;77(21);e104-e7.
Figures