1478
Automated Segmentation of Metastatic Lymph Nodes in FDG PET/MRI for Lymphoma Cancer Patients Using Multi-Scale Swin Transformers1Department of Circulation and Medical Imaging, NTNU, Trondheim, Norway, 2Department of Physics, NTNU, Trondheim, Norway, 3Department of Radiology and Nuclear Medicine, St. Olavs Hospital, Trondheim University Hospital, Trondheim, Norway
Synopsis
Keywords: Machine Learning/Artificial Intelligence, PET/MR, Automated Segmentation, Deep Learning, Transformers
Automated segmentation of tumors and designing a computer-assisted diagnosis system requires co-learning of imaging features from the complementary PET/MRI. We aim to develop an automated method for the segmentation of cancer-affected lymph nodes on Positron Emission Tomography/Magnetic Resonance Imaging (PET/MRI) using a modified Swin Transformer model (ST) integrated with a novel Multi-Scale Feature Fusion & Reorganization Module (MSFFRM). Our results with Hodgkin lymphoma (HL) and Non-Hodgkin lymphoma subtype Diffuse Large B-Cell Lymphoma (DLBCL) datasets show that our proposed model ST-MSFFRM achieved better performance in lesion segmentation in comparison to other state-of-the-art segmentation methods.Introduction
[18F]-Fluorodeoxyglucose (FDG) PET/MRI is frequently being used as an imaging modality for diagnosing cancer. For Hodgkin lymphoma (HL) and Non-Hodgkin lymphoma subtype Diffuse Large B-Cell Lymphoma (DLBCL) segmentation of cancer-affected lymph nodes are important for prognosis, staging, and response assessment of patients. However, making manual segmentations is time-consuming for patients with widespread metastatic disease.Automated segmentation of tumors and designing a computer-assisted diagnosis system requires co-learning of imaging features from the complementary PET/MRI. However, the small volume of the lymphoma lesions in comparison to the whole-body images adds complexity to the lesion segmentation. Therefore, in this work, we propose a model that captures both local and global features in multi-modal images. We designed an automated method for metastatic lymph nodes segmentation on PET/MRI of lymphoma patients using a modified Swin Transformer model (ST)1 integrated with a novel Multi-Scale Feature Fusion & Reorganization Module (MSFFRM).
Our technical contributions in this research work are twofold:
- We constructed a modified ST network having a Local Branch and a Global Branch to consider both global and local features from the multi-modal PET/MR images.
- We designed a novel MSFFRM to fuse the global and local features throughout multiple transformers.
Dataset
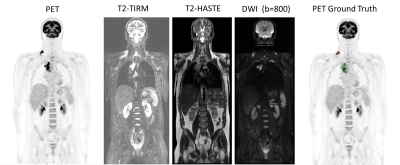
FDG PET/MRI baseline and interim images of 18 HL and 32 DLBCL lymphoma patients were analyzed and manually segmented using 3D Slicer and thereafter validated by an experienced nuclear medicine physician. Two different reader groups, each consisting of a radiologist and a nuclear medicine physician, have contributed to the clinical reading of the PET/MR images following standardized protocols. We designed a multi-channel input for our proposed architecture which, in addition to having the PET image as input, also uses multiparametric MRI such as T2-weighted HASTE MRI, T2 TIRM MRI, and Diffusion Weighted Image (DWI) with b = 800 s/mm2. Figure 3 presents the input images used for the model training and testing.Proposed Method
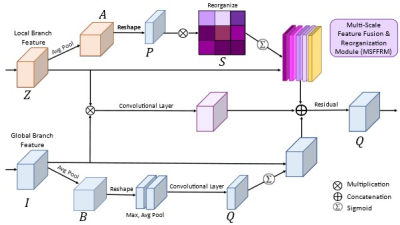
Inspired by Swin Unet 1, we combined CNN and superpixel 2 into a Local Branch for acquiring local-level features from input images. Global Branch follows the encoder architecture. The Global branch and local branch combined with the Self-Attention (SA) method in MSFFRM capture global dependencies in image feature learning and provide wide-ranging contextual information.Our modified ST consists of the encoder, decoder, bottleneck, and optimized skip connection using Multi-Scale Feature Fusion & Reorganization Module (MSFFRM) for improved training. Figure 1 illustrates the overall design of the proposed ST-MSFFRM model in detail. As an alternative to the conventional Swin Transformer block consisting of two identical Multi-head Self-Attention (MSA) modules, we used the Local Scale Multi-head Self-Attention (LS-MSA) module for the first Transformer followed by the Global Scale Multi-head Self-Attention (GS-MSA) module for the second Transformer3. In addition to the LS-MSA and GS-MSA, each ST block comprises of Layer Normalization (LN), residual connection, and 2-layer Multi-Layer Perceptron (MLP). Figure 2 depicts the detailed structure of our proposed Multi-Scale Feature Fusion & Reorganization Module (MSFFRM).
ST-MSFFRM was compared to three state-of-the-art segmentation methods: U-Net4, Swin Unet1, and D-Former3 using the clinical lymphoma dataset.
Implementation Details
The 64 PET/MRI examinations were divided into 85/15 ratio for training (N=53) and testing (N=11). Furthermore, multiple data augmentation techniques (random rotation, flip, noise, blur, and improved contrast) were performed for the training dataset, resultantly increasing the training dataset to 312 image sets. Our model is implemented in Python 3.6 and Pytorch 1.7.0. We trained our model on an NVIDIA Tesla P100 GPU with 16GB of memory. The weights pre-trained on ImageNet were used to initialize the model parameters for the ST-based models.Results
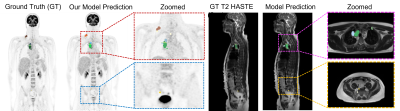
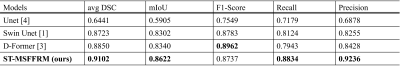
Our qualitative and quantitative results show that the model can segment cancer-affected lesions. Both a 4-fold and 13-fold cross-validation were performed for the training of the proposed model ST-MSFFRM. We used the average Dice-Similarity coefficient (DSC), mean Intersection over Union (IoU), F1-Score, Precision, and Recall as evaluation metrics to evaluate lesion segmentation. The segmentation results of our proposed model are visualized in Figure 4. Results from the testing of our model ST-MSFFRM with U-Net4, Swin Unet1, and D-Former3 in terms of average DSC and mIoU are shown in Figure 5. Our proposed model attained an average DSC of 0.9102 with 0.9236 precision.Discussion & Conclusion
Our ST-MSFFRM model automatically detected cancerous lymph nodes in the multi-modal (PET/MR) testing cohort. Performance evaluation of our model in comparison with other state-of-the-art models shows its potential for medical segmentation with a small dataset. Future research will focus on improving the dice score, as the model can be re-trained on larger datasets with improved normalization to achieve continuous training optimizations for better results. Furthermore, this method can be used on other datasets for the automatic segmentation of several types of cancer.Acknowledgements
This work was supported by the Trond Mohn Foundation for the 180°N (Norwegian Nuclear Medicine Consortium) project.References
[1] Cao, H., Wang, Y., Chen, J., Jiang, D., Zhang, X., Tian, Q., & Wang, M. (2021). Swin-unet: Unet-like pure transformer for medical image segmentation. arXiv preprint arXiv:2105.05537.
[2] Wang, Z., Min, X., Shi, F., Jin, R., Nawrin, S. S., Yu, I., & Nagatomi, R. (2022). SMESwin Unet: Merging CNN and Transformer for Medical Image Segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 517-526). Springer, Cham.
[3] Wu, Y., Liao, K., Chen, J., Wang, J., Chen, D. Z., Gao, H., & Wu, J. (2022). D-former: A u-shaped dilated transformer for 3d medical image segmentation. Neural Computing and Applications, 1-14.
[4] Ronneberger, O., Fischer, P., & Brox, T. (2015, October). U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention (pp. 234-241). Springer, Cham.
Figures