1477
Assessment of Breast Cancer Molecular Subtypes with a mMRI-based Feature Fusion Radiomics Model: Mimicking Radiologists’ Diagnostic Approach
Wanli Zhang1,2, Fangrong Liang1,2, Jiamin Li1,2, Yongzhou Xu3, Aaron Zhang3, Xinqing Jiang1,2, Xin Zhen4, and Ruimeng Yang1,2
1Department of Radiology, The Second Affifiliated Hospital, School of Medicine, South China University of Technology, Guangzhou, China, 2Department of Radiology, Guangzhou First People’s Hospital, Guangzhou, China, 3Philips Healthcare, Guangzhou, China, 4School of Biomedical Engineering, Southern Medical University, Guangzhou, China
1Department of Radiology, The Second Affifiliated Hospital, School of Medicine, South China University of Technology, Guangzhou, China, 2Department of Radiology, Guangzhou First People’s Hospital, Guangzhou, China, 3Philips Healthcare, Guangzhou, China, 4School of Biomedical Engineering, Southern Medical University, Guangzhou, China
Synopsis
Keywords: Breast, Radiomics, breast cancer, molecular receptor status, feature fusion
Since breast cancer is a highly heterogeneous tumor, prognosis and treatment response differ significantly according to different molecular subtypes. Based on multiparametric magnetic resonance imaging (MRI), we developed a feature fusion radiomics (RFF) model and investigated its performance in identifying the molecular receptor status of breast cancer. Mimicking the diagnostic approach of the radiologists by integrating image information from different MR sequences, the RFF model outperformed any single MRI-based radiomics model, demonstrating its potential for molecular subtypes classification of breast tumors.Introduction
Previous studies showed that radiomics has been successfully applied to preoperatively assess different molecular receptor statuses of breast cancer, predominantly by implementing one or two selected single MRI sequence-derived images, such as T2WI, DWI-derived ADC maps, and the early phase of dynamic contrast-enhanced (DCE) MRI1-3. However, on a daily work basis, radiologists must go through all acquired MRI sequences of patients, such as T1WIs, T2WIs, DWIs with various b values, ADC maps, and all phases of DCE MRIs, in order to make a final diagnosis. Based on mimicking the work pattern of a radiologist, we developed a novel feature fusion radiomics (RFF) model that could fuse radiomics features from multiparametric MRIs to determine its performance in distinguishing different statuses of molecular receptors in breast cancer preoperatively.methods
Data collectionFrom January 2017 to April 2022, 460 patients with 466 pathology-confirmed breast cancer who underwent breast MRI were retrospectively collected [luminal A: n=142; luminal B: n=194, human epidermal growth factor receptor 2 (HER2) enriched BC (HEBC): n=76; triple-negative BC (TNBC): n=54; among hormone receptor (HR) positive (HR+): n=238 and HR negative (HR-): n=99], as shown in Figure 1. All patients underwent the routine breast MRI examination on a 1.5T MR scanner (uMR 560, United Imaging) in Guangzhou First People’s Hospital, including T1WI, T2WI, DWIs (with three b values of b=0 s/mm2, 600 s/mm2, 800 s/mm2, the last two being denoted DWI600, DWI800, respectively), and six continuous DCE MRI (DCE1-6) scans with approximately 62 seconds per phase.
Tumor segmentation and radiomics feature selection
Ten volumes of interest (VOIs) for each lesion were delineated slice-by-slice on T2WI, DWI600, DWI800, ADC map, and DCE1-6 images. A total of 109 radiomics features were extracted from each VOI using Pyradiomics, an open-source software toolkit for radiomics analysis4, and then fed into 150 classification models that were constructed based on 10 classifiers and 15 feature selection methods in Table 1.
RFF model development and evaluation
Selecting ~72% cases (n=337) randomly, a random training cohort was composed to analyze all radiomics features of each sequence, analogous to the initial reviewing of a patient’s all MR images by a radiologist, determining the optimum sequence that can identify HR+ and HR- BC, TNBC and HEBC, as well as TNBC and non-TNBC. Multiple sequence feature fusion was then performed using radiomics features extracted from the top four high-performance sequences, similar to a final review by a radiologist focusing on sequences with specific features after a preliminary review, to identify the best combination of sequences to develop the RFF models respectively (Figure 2). Finally, all cases were randomly divided into a validation cohort (n=337) and a test cohort (n=129) at a ratio of 3:1, to verify the performance of the optimum single sequence and the fusion sequence. The RFF model was further evaluated via receiver operative characteristic (ROC) analysis and paired samples Wilcoxon signed rank test.
Results
By analyzing the dominant radiomics features of each sequence, the optimal discriminative sequence for discriminating HR+ vs. HR-, TNBC vs. HEBC, and TNBC vs. non-TNBC were DWI600, ADC and ADC, respectively, with the highest AUC of 0.787, 0.788, and 0.809 in the training cohort (Figure 3), and achieved the best AUC values of 0.767 and 0.768, 0.769 and 0.718, 0.784 and 0.735 in the validation cohort and test cohort, respectively. Afterward, the top four superior sequences for molecular subtype classification were selected to build the RFF model, demonstrating that the model RFF (DWI600+DWI800+DCE5), RFF (ADC+DCE2+DCE4) and RFF (ADC+DWI600+T2WI+DCE2) outperformed the single optimal sequence with an AUC of 0.778 and 0.726, 0.787 and 0.773, 0.818 and 0.773, respectively in the validation cohort and test cohort (both p<0.05 except HR+ vs. HR- ), as shown in Table 2.Discussion
Simulating the radiologist’s working pattern, we analyzed radiomics features from multiparameter MRI comprehensively, screening the most discriminative MRI sequences for distinguishing HR+ vs. HR -, TNBC vs. HEBC, and TNBC vs. non-TNBC, with the former being DWI600, and the latter both being ADC. Our study showed good performance with all AUC>0.7 and compensated for the limitation of previous researches of identifying different molecular receptor statuses in breast cancer1, 5, 6, which were employing only one or two MRI sequence(s) such as T2WI or ADC without exploring all routine multiparameter MRI sequences. In addition, we hypothesized that managing MRI sequences in a collaborative pattern would provide multi-dimensional image information, just as radiologists would concentrate on significant sequences after preliminary review. Incorporating class structure information, the feature fusion method proposed in the RFF framework has been proven to perform effective feature fusion by unitizing the constructed between-class matrix in our previous study7, 8. Thus, the fused features from different MR sequences were not only integrated but also more representative and discriminative, as our results showed that model RFF(DWI600+DWI800+DCE5), RFF (ADC+DCE2+DCE4) and RFF (ADC+DWI600+T2WI+DCE2) outperformed other MRI sequences in the identification of HR+ vs. HR -, TNBC vs. HEBC and TNBC vs. non-TNBC, respectively.Conclusion
The RFF model we developed could imitate the radiologists’ diagnostic approach to determine different molecular receptors of breast cancer preoperatively, which has great potential in the diagnosis or prognostic prediction of breast tumors.Acknowledgements
No acknowledgement found.References
1. Zhang Y, Chen J, Lin Y, et al. Prediction of breast cancer molecular subtypes on DCE-MRI using convolutional neural network with transfer learning between two centers. European Radiology, 2021,31(4):2559-2567.2. Qin Y, Tang C, Hu Q, et al. Assessment of Prognostic Factors and Molecular Subtypes of Breast Cancer With a Continuous-Time Random-Walk MR Diffusion Model: Using Whole Tumor Histogram Analysis. J Magn Reson Imaging, 2022.
3. Xie T, Wang Z, Zhao Q, et al. Machine Learning-Based Analysis of MR Multiparametric Radiomics for the Subtype Classification of Breast Cancer. Frontiers in Oncology, 2019,9.
4. van Griethuysen J, Fedorov A, Parmar C, et al. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res, 2017,77(21):e104-e107.
5. Lee J Y, Lee K, Seo B K, et al. Radiomic machine learning for predicting prognostic biomarkers and molecular subtypes of breast cancer using tumor heterogeneity and angiogenesis properties on MRI. European Radiology, 2021.
6. Leithner D, Bernard-Davila B, Martinez D F, et al. Radiomic Signatures Derived from Diffusion-Weighted Imaging for the Assessment of Breast Cancer Receptor Status and Molecular Subtypes. Molecular Imaging and Biology, 2020,22(2):453-461.
7. Haghighat M, Abdel-Mottaleb M, Alhalabi W. Discriminant Correlation Analysis: Real-Time Feature Level Fusion for Multimodal Biometric Recognition. IEEE transactions on information forensics and security, 2016,11(9):1984-1996.
8. Wu J, Liang F, Wei R, et al. A Multiparametric MR-Based RadioFusionOmics Model with Robust Capabilities of Differentiating Glioblastoma Multiforme from Solitary Brain Metastasis. Cancers (Basel), 2021,13(22).
Figures
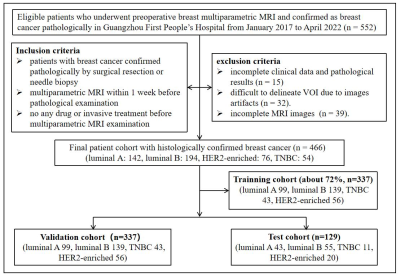
Figure 1. Flow chart of the study population with inclusion and exclusion criteria. HER2: human epidermal growth factor receptor 2. TNBC: triple-negative breast cancer.
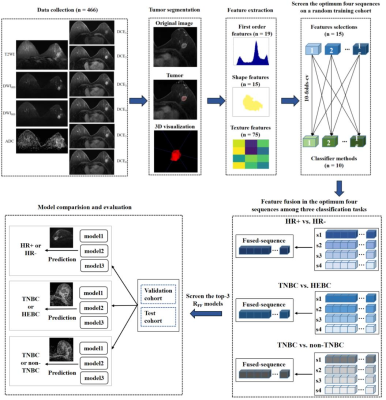
Figure 2. Flow chart of the study. HR: hormone receptor; TNBC: triple-negative breast cancer; HEBC: human epidermal growth factor receptor 2-enriched BC.
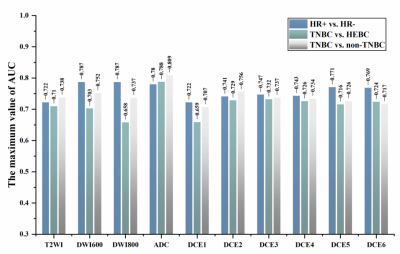
Figure 3. Optimal sequence with the maximum AUC value in identifying HR+ vs. HR- (DWI600), TNBC vs. HEBC (ADC), and TNBC vs. non-TNBC (ADC) in the training cohort. HEBC: human epidermal growth factor receptor 2-enriched BC;TNBC: triple-negative breast cancer. HR: hormone receptor.

Table 1. Feature selection methods and classifiers
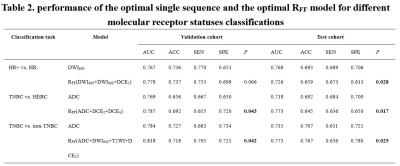
Table 2. performance of the optimal single sequence and the optimal RFF model for different molecular receptor status classifications. P value: compared the performance between the optimal single sequence and the optimal RFF model in the validation cohort and test cohort. Significant values (p < 0.05) are presented in bold. HR, hormone receptor; HEBC: human epidermal growth factor receptor 2-enriched BC; TNBC: triple-negative breast cancer. AUC, area under the receiver operative characteristic curve; SEN, sensitivity; SPE, specificity; ACC, accuracy.
DOI: https://doi.org/10.58530/2023/1477