1467
A Deep Learning-Based Tool for Analyzing the Female Reproductive System in MR images1Voxelwise Imaging Technology Inc., Vancouver, BC, Canada, 2Prenuvo, Vancouver, BC, Canada
Synopsis
Keywords: Uterus, Data Analysis
MRI is a powerful imaging technique for examining the anatomy of the female reproductive system. However, due to cost-related concerns, ease of access, acquisition time, and necessity of expert reviewers for MR images, ultrasound is the primary modality of choice. To mitigate some of these concerns, we developed an AI-driven tool comprising seven neural networks that segments the regions of interest for the whole uterus, uterine zones, ovaries, and further identifies common benign gynecological conditions. We evaluated our package on a large representative population of 2955 sagittal T2-weighted pelvic scans to obtain normative aging-curves for various regions of interest.Introduction
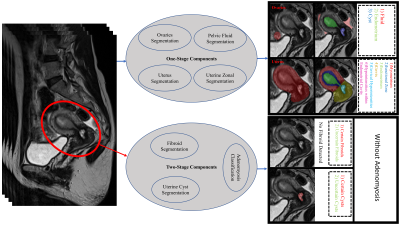
Although MR imaging gives substantial information on the uterus and ovaries, most of the existing AI-based methods for assessing the female reproductive system are developed for ultrasound1. Only lately, there have been limited publications on the use of AI in MRI of the uterus and ovaries, with the majority focusing on malignancies. However, benign gynecologic conditions are relatively common and can be accompanied by severe symptoms that may significantly influence a woman’s quality of life, highlighting the significance of developing AI-driven tools for monitoring their reproductive systems. The current literature of AI in MRI has already covered segmenting the uterus2,3, placenta4,5, endometrial cancer6,7,8,9, uterine tumors3 and further classifying sarcomas versus leiomyoma10,11,12, predicting outcome of fibroid embolization13 and ablation 14, classifying cervical cancer15, detecting ovarian cancer 16,17, and registering cervix images 18. Yet, to the best of our knowledge, no prior research has developed a comprehensive AI-driven tool for female reproductive system analysis from MR images. Our proposed package includes segmenting the uterus, uterine zones, fibroids, cysts, ovaries, pelvic fluid, and classifying adenomyosis (figure 1). Additionally, we derived the uterine and ovarian normative aging-curves by applying the proposed package on our large-scale dataset of sagittal T2-weighted sequences from a representative sample (N=2955) of adults ranging in age from 22 to 70.Material and Method
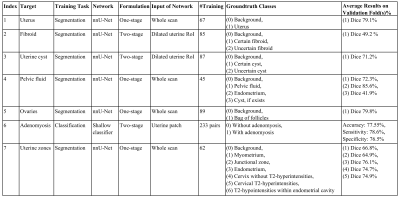
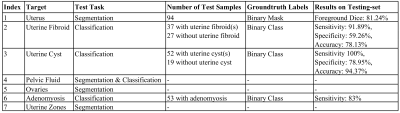
Data is collected from sagittal T2-weighted pelvic sequences obtained as part of a general preventive whole-body MRI screening program. We selected data for conditions based on delineations in our radiology reports. ITK-SNAP19 was used for annotating the ground-truth segmentation masks. Methods are formulated in either a one- or two-stage manner depending on the input of the network. Two-stage formulation improves efficiency while introducing errors from the uterus segmentation stage. For tasks formulated as semantic segmentation, nnU-Net20 framework was used. For classification tasks, we came up with a shallow-network developed in PyTorch Lightning21 with four convolutional blocks and one linear block. Further details regarding components are summarized in Table 1.Results and Discussion
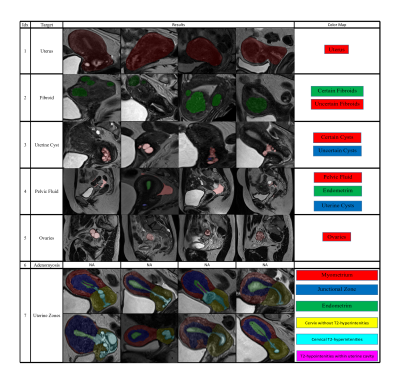
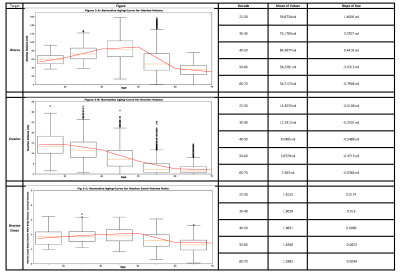
The components of our package are discussed, considering qualitative results (figure 2), results on large-scale dataset (figure 3), hold-out sets (table 2), and validation folds (table 1).1) Segmenting the uterus is essential for quantifying uterine volume towards monitoring its healthiness and age-related changes (figure 3-A). Additionally, the obtained mask reveals the uterine region of interest (RoI) for two-stage components.
2) Detecting fibroids from T2-weighted images is challenging owing to their diverse size and appearance, low contrast, and resemblance to other T2-hypointensities. Our method obtained a sensitivity of 91.89%, while specificity is 59.26% with majority of false-positives resulting from deficiency in distinguishing fibroids against other T2-hypointesities, mainly junctional zone (JZ). Employing false-positive T2-hypointensities as negative samples during training, multi-modal analysis, and formulating problem as instance segmentation [22] may further improve the specificity. Also, degenerated or calcified fibroids cause most false-negatives.
3) Uterine cyst model was trained on cases with Nabothian cysts, as indicated in our radiology reports. Nevertheless, the model segments various uterine cysts including those for cystic-adenomyosis and cesarean-scar, and achieves an accuracy of 94.37%. Localizing predicted cysts using zonal model can further determine their type.
4) Pelvic fluid: Training images were from patients delineated with fluid in Cul-de-Sac in our reports. However, reproductive-age women may have normal physiologic fluid. Thus, we could measure fluid volume whereas menopausal status and source of fluid are essential for declaring abnormal fluid in Cul-de-Sac condition. Identifying the source of fluid is possible through localizing fluid relative to potential sources e.g., ovaries or bowel.
5) Segmenting ovaries is required for determining volume and RoI of ovaries. Figure 3-B shows the normative aging-curve for ovarian volume, but counting follicles and identifying conditions were left for further development. Note that right and left ovaries are summed together.
6) Classifying adenomyosis was done using the shallow-network considering condition-related uterine texture changes. Balanced training data was collected in a pairwise manner, with each pair containing two uterus patches with and without adenomyosis. Despite obtaining a sensitivity of 83%, false-negative predictions were mostly characterized by borderline JZ thickening and minimal uterine texture change. Detecting JZ thickening and cystic-adenomyosis with zonal and uterine cyst models may further improve results.
7) Zonal Segmentation facilitates localization of uterine masses such as fibroid and cyst, along with measurement of zonal thicknesses, lengths, and volumes. Accordingly, a training-set of 62 samples from healthy and unhealthy patients of diverse ages was annotated with the five uterine zones, i.e., myometrium, JZ, endometrium, cervix, and cervical T2-hyperintensities. Further, a separate class for T2-hypointensities within the endometrial cavity was added. We proposed to calculate endometrial thickness as twice the maximum value in the 2D-distance transform applied to the 2D-slice of the endometrial binary mask with largest foreground area. Also, figure 3 depicts the normative aging-curve for the ratio of uterine main body volume to cervix for our large representative population compared with the curve for uterine and ovarian volume.
Conclusion
In this research, we developed a deep learning-based package for uterine and ovarian analysis, which shows the feasibility of AI-based MR image analysis tools to study the female reproductive system.Acknowledgements
We would like to thank the MRI Technologists, Patient Care, and Backend teams at Prenuvo for their contributions in data acquisition.References
1. Chen, Zhiyi, et al. "Artificial Intelligence in the Assessment of Female Reproductive Function Using Ultrasound: A Review." Journal of Ultrasound in Medicine 41.6 (2022): 1343-1353.
2. Kurata, Yasuhisa, et al. "Automatic segmentation of the uterus on MRI using a convolutional neural network." Computers in biology and medicine 114 (2019): 103438.
3. Zhang, Chen, et al. "HIFUNet: multi-class segmentation of uterine regions from MR images using global convolutional networks for HIFU surgery planning." IEEE Transactions on Medical Imaging 39.11 (2020): 3309-3320.
4. Twickler, Diane M., et al. "228: Automated segmentation of the human placenta and uterus with MR imaging using artificial intelligence (AI)." American Journal of Obstetrics & Gynecology 222.1 (2020): S158-S159.
5. Shahedi, Maysam, et al. "Deep learning-based segmentation of the placenta and uterus on MR images." Journal of Medical Imaging 8.5 (2021): 054001.
6. Hodneland, Erlend, et al. "Automated segmentation of endometrial cancer on MR images using deep learning." Scientific reports 11.1 (2021): 1-8.
7. Dong, Hsiang-Chun, et al. "Using deep learning with convolutional neural network approach to identify the invasion depth of endometrial cancer in myometrium using MR images: a pilot study." International Journal of Environmental Research and Public Health 17.16 (2020): 5993.
8. Urushibara, Aiko, et al. "The efficacy of deep learning models in the diagnosis of endometrial cancer using MRI: a comparison with radiologists." BMC Medical Imaging 22.1 (2022): 1-14.
9. Mao, Wei, et al. "A deep learning-based automatic staging method for early endometrial cancer on MRI images." Frontiers in Physiology 13 (2022).
10. Malek, Mahrooz, et al. "A diagnostic algorithm using multi-parametric MRI to differentiate benign from malignant myometrial tumors: Machine-Learning method." Scientific Reports 10.1 (2020): 1-12.
11. Dai, Mengying, et al. "Combining multiparametric MRI features-based transfer learning and clinical parameters: application of machine learning for the differentiation of uterine sarcomas from atypical leiomyomas." European Radiology (2022): 1-10.
12. Malek, Mahrooz, et al. "A machine learning approach for distinguishing uterine sarcoma from leiomyomas based on perfusion weighted MRI parameters." European journal of radiology 110 (2019): 203-211.
13. Luo, Yong-Heng, et al. "Deep learning based on MR imaging for predicting outcome of uterine fibroid embolization." Journal of Vascular and Interventional Radiology 31.6 (2020): 1010-1017.
14. Zheng, Yineng, et al. "Prediction of Clinical Outcome for High-Intensity Focused Ultrasound Ablation of Uterine Leiomyomas Using Multiparametric MRI Radiomics-Based Machine Leaning Model." Frontiers in Oncology 11 (2021).
15. Urushibara, Aiko, et al. "Diagnosing uterine cervical cancer on a single T2-weighted image: comparison between deep learning versus radiologists." European Journal of Radiology 135 (2021): 109471.
16. Xu, He-Li, et al. "Artificial intelligence performance in image-based ovarian cancer identification: A systematic review and meta-analysis." EClinicalMedicine 53 (2022): 101662.
17. Xu, Chen, et al. "Advances of Artificial Intelligence Application in Medical Imaging of Ovarian Cancers." Chinese Medical Sciences Journal 36.3 (2021): 196-203.
18. Guo, Peng, et al. "Unsupervised Deep Learning Registration of Uterine Cervix Sequence Images." Cancers 14.10 (2022): 2401.
19. Yushkevich, Paul A., et al. "User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability." Neuroimage 31.3 (2006): 1116-1128.
20. Isensee, Fabian, et al. "nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation." Nature methods 18.2 (2021): 203-211.
21. https://github.com/PyTorchLightning/pytorch-lightning[22] Finn, Chelsea, Pieter Abbeel, and Sergey Levine. "Model-agnostic meta-learning for fast adaptation of deep networks." International conference on machine learning. PMLR, 2017.
22. Baumgartner, Michael, et al. "nndetection: A self-configuring method for medical object detection." International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2021.
23. Kelsey TW, Dodwell SK, Wilkinson AG, et al. Ovarian volume throughout life: a validated normative model. PLoS One. 2013;8(9):e71465. Published 2013 Sep 3. doi:10.1371/journal.pone.0071465
Figures