1441
Differentiation between myelodysplastic syndrome and aplastic anemia using transfer learning of vision transformer
Miyuki Takasu1, Yasutaka Baba2, Konagi Takeda1, Saki Kawai1, Hiroaki Sakane1, Nobuko Tanitame1, Akihisa Tamura1, Makoto Iida1, and Kazuo Awai3
1Diagnostic Radiology, Hiroshima City Hiroshima Citizens Hospital, Hiroshima, Japan, 2Diagnostic Radiology, International Medical Center, Saitama Medical University, Hidaka, Japan, 3Hiroshima University Hospital, Hiroshima, Japan
1Diagnostic Radiology, Hiroshima City Hiroshima Citizens Hospital, Hiroshima, Japan, 2Diagnostic Radiology, International Medical Center, Saitama Medical University, Hidaka, Japan, 3Hiroshima University Hospital, Hiroshima, Japan
Synopsis
Keywords: Bone, Machine Learning/Artificial Intelligence
We examined the use of a vision transformer (ViT)-based deep learning model for the task of differentiation between aplastic anemia and myelodysplastic syndrome using lumbar T1-weighted images. Three sagittal images per patient were obtained and made square using zero-padding and were resized (224 × 224). The overall accuracy and area under the curve of the pre-trained ViT model were higher than those of ViT without pre-training, ResNet-110, and BinaryNet at the optimum hyperparameters. We utilized Grad-CAM images to highlight the information that is important for decision-making. ViT combined with Grad-CAM successfully recognized variability in the distribution of bone marrow components.Introduction
Aplastic anemia (AA) is characterized by various degree of pancytopenia and hypocellular bone marrow (BM) .1 Allogeneic hematopoietic stem cell transplantation (HSCT) is a curative option for AA, with a reported possibility of long-term cure of 75%-80%.2 Myelodysplastic syndrome (MDS) is a clonal disease of hematopoietic cells, characterized by ineffective hematopoiesis in the BM and a high risk of transformation into acute myeloid leukemia (AML).3 Allogeneic HSCT is the only curative treatment for MDS, although relapse remains the primary cause of mortality after receiving allogeneic HSCT.4 The etiologies of MDS and AA are distinct; however, differentiation between AA and MDS can be challenging because the clinical and morphologic differences in BM might be subtle.In recent years, the rapid development of deep learning technology has enabled various types of image processing, including that of MR images. Deep learning technology is largely based on convolutional neural networks (CNNs). Recently, however, CNNs have been out-performed by attention-based architecture, known as transformers.5
In this study, we examined the use of a vision transformer (ViT)-based deep learning model for the task of differentiation between AA and MDS using lumbar T1-weighted images.
Methods
This retrospective, multi-institution study was approved by the Institutional Review Board of Hiroshima University Hospital and the five participating centers.Patients with a pathologically confirmed diagnosis of MDS (n = 70) or AA (n = 84) underwent MRI performed using the spine protocols used at each institution, which included 2D sagittal T1-weighted spin-echo sequences using either a 3-T system (n = 120) or a 1.5-T system (n = 34).
For the lumbar T1-weighted images, we used three images (mid-sagittal and bilateral para-mid-sagittal images) per patient. Images were made square using zero-padding and resized (224×224). We created a single dataset obtained from all six institutions and used 10-fold cross-validation for training the algorithms. We then tested the models on data from the two or three most-recent patients from each institution.
Four training algorithms were implemented: (1) ViT, a fine-tuned version of ImageNet pre-trained data-efficient image transformers (DeiT) as the base model, (2) ViT without pre-training, using the same model architecture as ViT, (3) ResNet-110, and (4) BinaryNet.
Hyperparameters for training were optimized by evaluating the performance of the validation process. The batch size was 16 and data augmentation included random rotation (± 25°), horizontal flipping, vertical flipping, color jittering, and random erasing (p = 0.2). Networks were trained for 100 epochs using the PyTorch machine learning framework. The Adam and Nesterov algorithms were used as optimizers for ViT and CNN models, respectively.
Measures of precision, F1-score, sensitivity, specificity, area under the curve (AUC), and overall accuracy were estimated and compared between the four algorithms. Gradient-weighted Class Activation Mapping (Grad-CAM) was used to visualize the regions of MR images that contributed to the models’ decision-making.
Results
Table 1 summarizes the precision, F1-score, sensitivity, specificity, AUC, and overall accuracy for differentiation between MDS and AA using T1-weighted image datasets. The overall accuracy of the pre-trained ViT model was 91.3%, which was higher than that of ViT without pre-training (65.2%), ResNet-110 (87.0%), and BinaryNet (65.2%) at the optimum hyperparameters. Confusion matrices for the models are provided in Figure 1.For differentiation between the two entities, the ViT model achieved an AUC of 0.909, which was higher than that of ResNet-110 CNN (AUC of 0.886). Without pre-training, the ViT model was inferior to ResNet-110 and BinaryNet CNNs, with an AUC of 0.609.
Grad-CAM images served as a reference for distribution of fatty and cellular marrow (Figure 2). The focus of ViT was mainly on cellular marrow (i. e., low signal intensity) for relatively homogeneous BM. For BM with foci of cellular marrow, Grad-CAM tended to focus on high signal fatty marrow.
Discussion
We demonstrated the utility of ViT for distinguishing between patients with MDS and AA using MR images of lumbar BM.ViT is a model of a transformer that has been extended for use in image classification in the medical field. Pre-training ViT on a larger dataset and subsequently transferring the learning to a smaller dataset has out-performed the CNN architectures in tasks of image classification. 5 In this study, the overall accuracy and AUC of the pre-trained ViT model were higher than those of ResNet-110 and BinaryNet, and these results were in agreement with those of previous studies. 5
ViT combined Grad-CAM images showed that ViT recognized different distributions of fatty marrow, hematopoietic marrow, and ineffective intramedullary erythropoiesis, and differentiated MDS from AA. However, it might be difficult to extract a specific pattern for these entities from BM with scattered foci of cellular marrow. ViT-assisted differentiation of these entities requires further investigation.
Conclusion
The results demonstrated that the ViT model accurately differentiated between AA and MDS using lumbar T1-weighted images, which are routinely acquired in medical practice, and shows potential for the non-invasive diagnosis of these entities. This study also demonstrated the efficacy of the ViT model over CNN-based methods. We utilized explainability-driven Grad-CAM images to highlight the information that is important for decision-making.Further investigation of ViT implementations with larger patient cohorts and in combination with sequences such as diffusion-weighted imaging could improve the performance of the model.
Acknowledgements
The authors thank Takashi Abe, MD, for implementing ViT and the insightful discussions.References
- Thakur W, Anwar N, Samad S, et al. Assessment of Hepatic Profile in Acquired Aplastic Anemia: An Experience From Pakistan. Cureus. 2022;14(9): e29079.
- Zhu Y, Gao Q, Hu J, et al. Allo-HSCT Compared with Immunosuppressive therapy for acquired aplastic anemia: A system review and meta-analysis. BMC Immunol. 2020;21(1):10.
- Vittayawacharin P, Kongtim P, Ciurea SO. Allogeneic stem cell transplantation for patients with myelodysplastic syndromes. Am J Hematol. 2022 Oct 17. doi: 10.1002/ajh.26763. Online ahead of print.
- Piemontese S, Boumendil A, Labopin M. Leukemia relapse following unmanipulated haploidentical transplantation: A risk factor analysis on behalf of the ALWP of the EBMT. J Hematol Oncol. 2019;12:1.
- Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, et al. An image is worth 16x16 words: Transformers for image recognition at scale. https://openreview.net/pdf?id=YicbFdNTTy. 2021.
Figures
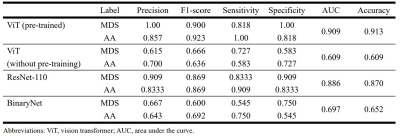
Table 1. Comparison of performance
of the various deep learning algorithms for differentiation between aplastic
anemia (AA) and myelodysplastic syndrome (MDS).
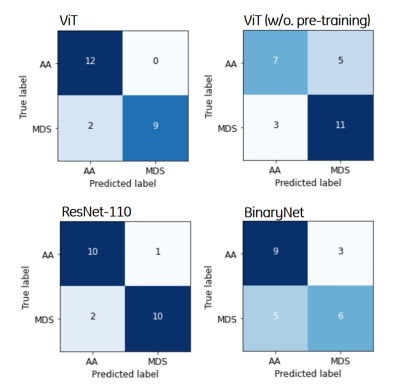
Figure 1. Confusion matrices for
differentiating between aplastic anemia and myelodysplastic syndrome of the
four deep learning algorithms. The vertical and horizontal axes in the figures
consist of the ground truth and predicted labels, respectively.
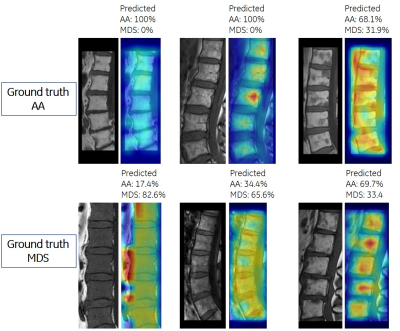
Figure 2. Visualization of
critical factors in decision making for differentiation between aplastic anemia
and myelodysplastic syndrome by vision transformer. In each subfigure, the left-sided
image presents the input to vision transformer, and the right-sided figure
presents the predicted probabilities for each class and highlights the factors
critically corresponding to the predicted class.
DOI: https://doi.org/10.58530/2023/1441