1396
Optimization of structural connectomes and scaled patterns of structural-functional decoupling in Parkinson’s disease1Clinical Medical College, Yangzhou University, Yangzhou, China, 2GE Healthcare, MR Research China, Beijing, China
Synopsis
Keywords: Parkinson's Disease, Neurodegeneration
Parkinson’s disease (PD) is manifested with disrupted topology of structural connection network (SCN) and functional connection network (FCN). However, SCN and its interactions with FCN remain to be further investigated. This multimodality study attempted to precisely characterize the SCN using diffusion kurtosis imaging (DKI) and further identify the neuropathological pattern of SCN-FCN decoupling. Our study verified the optimization of SCN by applying DKI metrics and its implications for the clinical diagnosis of PD. Moreover, SCN-FCN decoupling is consequent from mismatched disruptions of the SCN and FCN, identifying pathophysiological neuroimaging features for disturbed neural circuits in PD.
Introduction
Parkinson’s disease (PD) is conceptualized as neuropathological disconnection syndrome, characterized by the presence of broad disturbances among neural circuits1. Recent magnetic resonance imaging (MRI) studies constructed a brain connectome via anatomical fibers traced by diffusion tensor imaging (DTI) or neuronal coactivation mapped from blood oxygen level-dependent (BOLD). However, several prominent inconsistencies were also encountered when comparing structural and functional networks2-4. One possible interpretation would be a vulnerable integrity agreement between the structural connection network (SCN) and functional connection network (FCN) in neurodegenerative conditions. Nevertheless, SCN-FCN decoupling in the neurodegenerative processes of PD remains to be confirmed and characterized in vivo.Additionally, a major issue of DTI in the construction of structural networks was an inadequate tracing of axons and an insensitivity to the isotropic environment of neurons5. To address this, diffusion kurtosis imaging (DKI) was proposed as an advanced alternative for the detection of diffusion profiles and applied to quantify the deviated fiber tracts and subcortical nuclei in PD. The current work thus sought to analyze the network topology and diagnostic ability based on connection strength by constructing both SCNs and FCNs in patients with PD by comparing with healthy controls (HCs). Moreover, the potential patterns of SCN-FCN decoupling at global, nodal and modular levels in PD were explored by following the quantification of SCN-FCN coupling.
Materials and Methods
SubjectsA final sample consisted of 69 (30 male and 39 female) patients with PD, and 70 (34 male and 36 female) matched HCs were recruited. The Unified Parkinson’s Disease Rating Scale part III (UPDRS-III) and Hoehn-Yahr scale were scored for disease severity and stage of PD. The global cognition evaluation was performed using the Mini-Mental State Examination (MMSE) and Montreal Cognitive Assessment (MoCA) for all participants.
MRI experiment
MRI experiments were performed using a 3.0-tesla MRI scanner (Discovery MR750, GE, USA) with an 8-channel phased array head coil. Resting-state functional MRI data of the whole brain were acquired as follows: TR, 2,000 ms; TE, 30 ms; FA, 90°; slice thickness, 4 mm without gap; FOV, 240 × 240 mm2; matrix size, 64 × 64; voxel size, 4.0 × 4.0 × 4.0 mm3; 240 time points; total scan time, 480 s. DKI images were acquired using single-shot spin-echo echo-planar imaging with the following parameters: TR, 5800 ms; TE, 100 ms; FOV, 240 × 240 mm2; matrix size, 100 × 100; voxel size, 2.4 × 2.4 × 2.4 mm3; b-values, 0; 1250 and 2500 s/mm2; diffusion gradient directions, 15 per b value; number of slices, 35; and total scan time: 330 seconds.
Data analysis
Functional data were preprocessed in SPM 12 embedded in the MATLAB 2018a platform. DKI data were preprocessed using MATLAB software and Diffusion Kurtosis Estimator. The networks construction and topology analysis were performed using the GRETNA toolbox. The quantification of SCN-FCN coupling at three levels (global, nodal and modular) were conducted by calculating the correlation coefficients32,33.
Statistical analysis
Classification performance of connection strengths was analyzed by using least absolute shrinkage and selection operator for feature selection and support vector machine approach for the classification. For FCN and SCN, the topological properties and SCN-FCN coupling coefficients were compared between groups using a two-sample t test in the GRETNA toolbox. The exploration of potential correlations between topological metrics and clinical scale scores in PD group was conducted using Spearman rank correlation test with SPSS 19.0 software. The threshold for statistical significance was set to a P value < 0.05.
Results
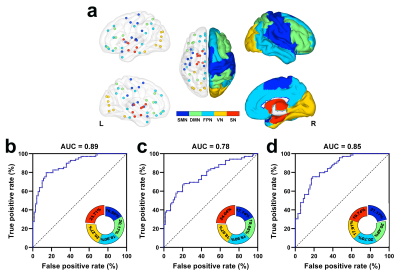
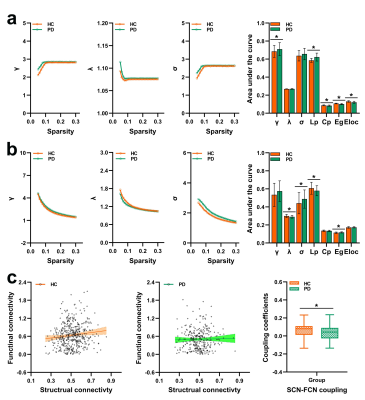
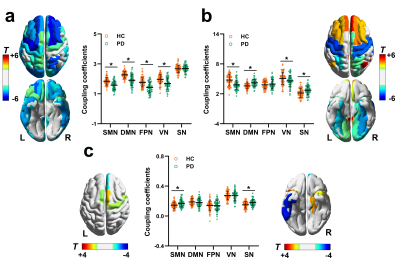
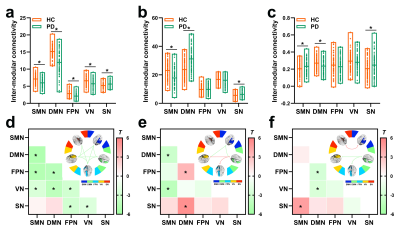
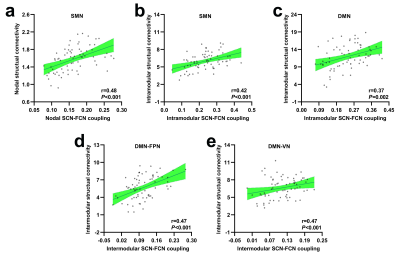
The SCN constructed by kurtosis metrics achieved optimal classification performance (area under the curve 0.89, accuracy 80.55%, sensitivity 78.40%, and specificity 80.65%, Fig.1). Along with diverse alterations of structural and functional network topology, the PD group exhibited decoupling across scales including: reduced global coupling (Fig.2); increased nodal coupling (Fig.3) within the sensorimotor network (SMN) and subcortical network (SN); higher intramodular coupling within the SMN and SN and lower intramodular coupling of the default mode network (DMN); decreased coupling between modules of DMN-fronto-parietal network and DMN-visual network, but increased coupling between the SMN-SN module (Fig.4). Several associations between the coupling coefficient and topological properties of the SCN, as well as between network values and clinical scores (UPDRS-III, MMSE and MoCA), were observed (Fig.5).Discussion and conclusion
In the present work, we constructed a microstructural network by employing DKI and further systemically identified the patterns of disconnection syndrome in patients with PD from the perspective of both SCN and FCN topology as well as their decoupling. Our study confirmed that kurtosis-derived SCN is optimal for exploration of topological alterations with an improved classification performance, and patients with PD suffered with neuropathological SCN-FCN decoupling at global, nodal and modular levels, serving as a complementary elucidation for disconnection syndrome.In conclusion, patients with PD encounter SCN-FCN decoupling across scales as the consequence of mismatched disruptions of the structural and functional connectome, which could be indicative of pathophysiological features for disturbed neural circuits. Hence, the current neuroimaging study provided empirical evidence for the comprehensive understanding of deficient information communications and underlying neurodegenerative mechanisms of PD.
Acknowledgements
No acknowledgement found.References
1. Borghammer P. The alpha-Synuclein Origin and Connectome Model (SOC Model) of Parkinson's Disease: Explaining Motor Asymmetry, Non-Motor Phenotypes, and Cognitive Decline. J Parkinsons Dis 2021;11:455-474.
2. Galantucci S, Agosta F, Stefanova E et al. Structural Brain Connectome and Cognitive Impairment in Parkinson Disease. Radiology 2017;283:515-525.
3. Baggio HC, Sala-Llonch R, Segura B et al. Functional brain networks and cognitive deficits in Parkinson's disease. Hum Brain Mapp 2014;35:4620-4634.
4. Aracil-Bolanos I, Sampedro F, Marin-Lahoz J et al. A divergent breakdown of neurocognitive networks in Parkinson's Disease mild cognitive impairment. Hum Brain Mapp 2019;40:3233-3242.
5. Kamagata K, Tomiyama H, Hatano T et al. A preliminary diffusional kurtosis imaging study of Parkinson disease: comparison with conventional diffusion tensor imaging. Neuroradiology 2014;56:251-258.
Figures