1395
Robustness of Autoencoder-based Classifier for fMRI-based Optimization of Single-sided Deep Brain Stimulation1GE Global Research, Niskayuna, NY, United States, 2University Health Network and University of Toronto, Toronto, ON, Canada
Synopsis
Keywords: Parkinson's Disease, fMRI
Successful treatment of Parkinson’s disease using deep brain stimulation (DBS) of the sub-thalamic nucleus (STN) requires an optimal set of DBS parameters that involves time-consuming programming sessions (~1 year) by the current standard-of-care optimization protocol. Functional magnetic resonance imaging (fMRI) and deep learning with autoencoder-based feature extraction from DBS-fMRI responses have provided a way to rapidly optimize the DBS parameters. In this work, we examine the robustness of the unsupervised autoencoder-based feature extraction method to changes in the activation patterns of the DBS-fMRI responses, which may be caused by patient motion, difference in stimulation side and disease condition.Introduction
Successful treatment of Parkinson’s disease (PD) using deep brain stimulation (DBS) of the sub-thalamic nucleus (STN) requires the optimization of stimulation parameters such as signal frequency, voltage, pulse width and contact location, which usually require time consuming programming sessions (~1 year) based on the current standard-of-care parameter optimization method1,2. Previously established functional magnetic resonance imaging (fMRI) and machine learning-assisted left-sided DBS parameter optimization for PD treatment has provided a way to rapidly classify DBS parameters as either optimal (patient clinical benefits are maximized and adverse effects are minimized) or non-optimal using features that were extracted from 16 pre-defined parcels in brain regions of motor function relevance3. Compared to the parcel-based feature selection method, an autoencoder (AE)-based autonomous feature extraction from DBS-fMRI response maps has been shown to improve the classification accuracy of fMRI-based single-sided DBS optimization, as the extracted features capture the neuro-functional patterns of each patient's DBS response4. The unique DBS-fMRI response maps at optimal or non-optimal parameters for PD patients can show changes in the activation and deactivation patterns due to patient motion, differences in disease condition or changes in the stimulation side. However, the ability of an AE model, trained on left-sided (or nominal) DBS-fMRI data, to extract physiologically meaningful features from response data that contain the aforementioned changes in the response pattern has not been assessed. Here, we implement an AE-based multilayer perceptron (MLP) model (AE-MLP) for fMRI-based classification of DBS parameters for PD patients that received left-sided DBS treatments, with the aim of investigating the robustness of the AE feature extraction model (trained on left-sided data only) to changes in the activation patterns of the input response maps.Materials and Methods
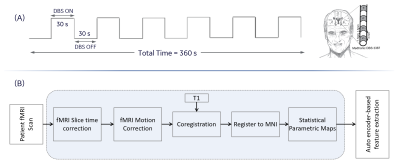
Our previously acquired 122 blood oxygenated level dependent fMRI data from 39 a priori clinically optimized PD patients (mean age$$$=$$$62.4$$$\pm$$$7.1, 20 males, 19 females) at 3.0 T were used in this work3.Single subject fMRI analyses: DBS-fMRI data from each of the 39 PD patients that have undergone left-sided DBS treatment was slice time corrected, motion corrected, rigidly registered to a T1-weighted image, non-linearly registered to a standard space Montreal Neurological Institute (MNI) brain, and spatially smoothed using a Gaussian kernel with a 6 mm full width at half maximum. The Art toolbox was used to detect and remove volumes with motion greater than 2 mm from the fMRI timeseries5. Statistical parametric maps (t-maps) were estimated from the preprocessed fMRI data using the designed 30-second DBS-ON/OFF paradigm. Other details of the processing steps are summarized in Figure 1. All data was processed using SPM12 (http://www.fil.ion.ucl.ac.uk) and MATLAB (Mathworks$$$-$$$Natick, MA, USA). To mimic changes that can occur in DBS-fMRI responses as a result of differences in stimulation side, disease condition and patient motion, the obtained response maps were passed through a left-right flip operation to displace the activated and deactivated regions horizontally.
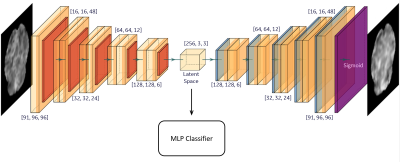
Model training and analyses: The implemented AE-MLP network consists of two stages. The first stage is automatic feature learning, where the AE network learns to map the high dimensional left-sided DBS-fMRI responses into latent feature vectors with low dimensionality through an encoder sub-network, and then a decoder sub-network learns to reconstruct the original inputs from their latent vectors. In the second stage, the latent feature vectors (length$$$=$$$2304) from the nominal and flipped response maps are normalized and separately fed into the AE-MLP model (trained on left DBS-fMRI responses) for optimal/non-optimal classification of the DBS parameter setting (Figure 2). The accuracy of the trained A-MLP model was estimated using nominal and flipped response maps via a 5-fold cross validation. The distribution of the latent vectors extracted from the nominal and flipped responses were compared using violin plots and cosine similarity index (CSI).
Results and Discussions
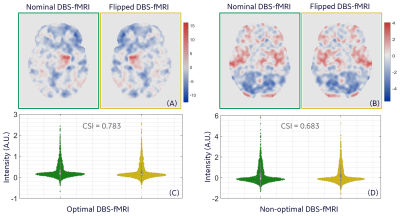
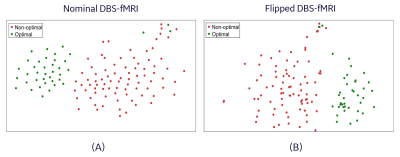
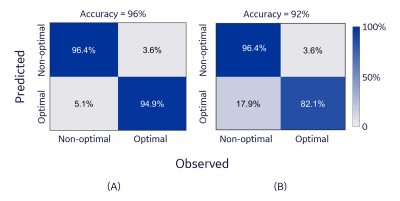
Representative nominal and flipped DBS-fMRI response maps, as well as the distribution of the extracted features for optimal and non-optimal DBS are shown in Figure 3. Despite the difference in the nominal and flipped response maps, the distributions of their respective AE-extracted features were similar with CSI values as high as 0.783 and 0.683 for the optimal and non-optimal response maps respectively. The t-distributed stochastic neighbor embedding (t-SNE) visualization of the extracted latent vectors for the entire patient cohort were clustered in a neuro-functionally meaningful manner for the nominal or flipped DBS-fMRI response maps (Figure 4). Since the extracted features of the nominal and flipped responses are comparable, the accuracy of the AE-MLP classification model for both datasets was also similar with a difference of 4% (Figure 5). Though our left-right flipping operation does not exactly capture the real-world differences that will be imposed on a DBS-fMRI acquisition by difference in stimulation side, disease condition and patient motion etc., these results indicate that the AE-based autonomous feature extraction method may be robust to differences in response maps that change the activated and deactivated regions horizontally. Our results also suggests that the left-right flipping operation may be used for data augmentation of single sided DBS-fMRI data, which is potentially useful for training more robust deep learning models.Conclusion
The AE-based feature extraction is robust to subtle differences in the activated regions of DBS-fMRI response maps obtained during fMRI-based DBS parameter optimization in PD patients.Acknowledgements
This work was supported by the Michael J. Fox foundation (grant number MJFF-008877, 2019).References
1. Lozano, A. M. et al. Deep brain stimulation: current challenges and future directions. Nat Rev Neurol 15, 148–160 (2019).
2. Picillo, M., Lozano, A. M., Kou, N., Puppi Munhoz, R. & Fasano, A. Programming Deep Brain Stimulation for Parkinson’s Disease: The Toronto Western Hospital Algorithms. Brain Stimulation 9, 425–437 (2016).
3. Boutet, A. et al. Predicting optimal deep brain stimulation parameters for Parkinson’s disease using functional MRI and machine learning. Nat Commun 12, 3043 (2021).
4. Ajala, A. et al. Autoencoder-Based Deep Learning Classifier for Deep Brain Stimulation Parameter Settings by fMRI. in OHBM 2022 Annual Proceedings (2022).
5. Mazaika, P. K., Hoeft, F., Glover, G. H., Reiss, A. L., & others. Methods and software for fMRI analysis of clinical subjects. Neuroimage 47, S58 (2009).
Figures